You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003674_01576
You are here: Home > Sequence: MGYG000003674_01576
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp900772575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp900772575 | |||||||||||
| CAZyme ID | MGYG000003674_01576 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1883; End: 4627 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 17 | 456 | 4.4e-71 | 0.49069148936170215 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.10e-20 | 23 | 515 | 14 | 486 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.22e-14 | 20 | 315 | 11 | 296 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 1.75e-09 | 24 | 164 | 4 | 154 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK09525 | lacZ | 2.46e-07 | 127 | 438 | 165 | 481 | beta-galactosidase. |
| pfam00703 | Glyco_hydro_2 | 8.53e-07 | 192 | 295 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO25184.1 | 0.0 | 19 | 913 | 19 | 923 |
| QCQ49396.1 | 0.0 | 16 | 913 | 29 | 922 |
| QRM69893.1 | 0.0 | 16 | 913 | 29 | 922 |
| AUI45420.1 | 0.0 | 16 | 913 | 29 | 922 |
| QTO27419.1 | 0.0 | 16 | 913 | 29 | 922 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D1P_A | 1.50e-13 | 23 | 440 | 41 | 455 | Apostructure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii],6D1P_B Apo structure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii] |
| 6JZ1_A | 5.56e-12 | 23 | 547 | 19 | 571 | Apostructure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ1_B Apo structure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ5_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ5_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ6_A b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ6_B b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_A b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_B b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ8_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus],6JZ8_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus] |
| 5Z18_A | 5.76e-12 | 23 | 547 | 43 | 595 | Thecrystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_B The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_C The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_D The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_E The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_F The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z19_A The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_B The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_C The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_D The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_E The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_F The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],6JZ2_A b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution [[Ruminococcus] gnavus],6JZ2_B b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution [[Ruminococcus] gnavus],6JZ3_A b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin [[Ruminococcus] gnavus],6JZ3_B b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin [[Ruminococcus] gnavus],6JZ4_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam [[Ruminococcus] gnavus],6JZ4_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam [[Ruminococcus] gnavus] |
| 6HPD_A | 3.89e-11 | 52 | 483 | 65 | 513 | Thestructure of a beta-glucuronidase from glycoside hydrolase family 2 [Formosa agariphila KMM 3901] |
| 1YQ2_A | 3.56e-08 | 15 | 419 | 27 | 444 | ChainA, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_B Chain B, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_C Chain C, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_D Chain D, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_E Chain E, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_F Chain F, beta-galactosidase [Arthrobacter sp. C2-2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P06760 | 2.46e-11 | 18 | 470 | 35 | 504 | Beta-glucuronidase OS=Rattus norvegicus OX=10116 GN=Gusb PE=1 SV=1 |
| T2KN75 | 2.12e-10 | 52 | 483 | 56 | 504 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| O97524 | 3.48e-09 | 16 | 568 | 33 | 621 | Beta-glucuronidase OS=Felis catus OX=9685 GN=GUSB PE=1 SV=1 |
| P26257 | 3.29e-08 | 260 | 438 | 232 | 409 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| P05804 | 8.04e-07 | 24 | 315 | 15 | 292 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
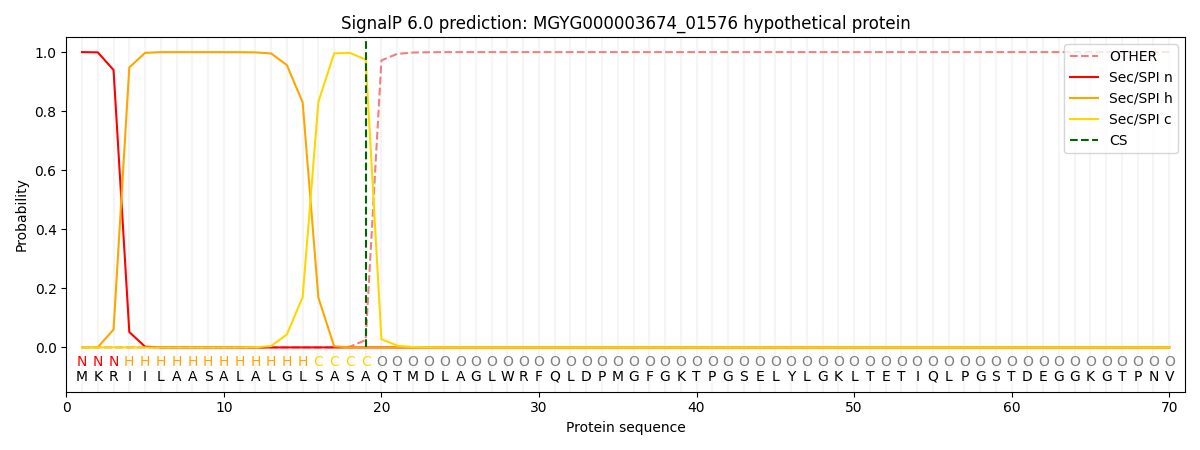
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000305 | 0.998973 | 0.000185 | 0.000179 | 0.000165 | 0.000154 |
