You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003681_00850
You are here: Home > Sequence: MGYG000003681_00850
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides stercoris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides stercoris | |||||||||||
| CAZyme ID | MGYG000003681_00850 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 107746; End: 110019 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 490 | 752 | 1.8e-48 | 0.6798679867986799 |
| GH10 | 60 | 179 | 5.6e-19 | 0.37623762376237624 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.90e-34 | 490 | 750 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 5.18e-33 | 490 | 752 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.91e-24 | 488 | 757 | 117 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 5.98e-13 | 63 | 151 | 11 | 98 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.58e-09 | 87 | 179 | 58 | 147 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADD61487.1 | 0.0 | 1 | 757 | 1 | 757 |
| ADD61481.1 | 0.0 | 1 | 757 | 1 | 757 |
| QUT46089.1 | 2.02e-308 | 1 | 757 | 1 | 719 |
| QRQ48288.1 | 2.86e-308 | 1 | 757 | 1 | 719 |
| ANU56191.1 | 6.47e-300 | 9 | 754 | 2 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGQ_A | 4.12e-22 | 142 | 294 | 4 | 146 | PbXyn10CCBM APO [Prevotella bryantii B14] |
| 2F8Q_A | 2.67e-17 | 451 | 747 | 83 | 348 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 2FGL_A | 2.70e-17 | 451 | 747 | 84 | 349 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2FGL_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 5EFD_A | 2.73e-17 | 451 | 747 | 85 | 350 | Crystalstructure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5EFD_B Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5XC0_A Crystal structure of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5XC0_B Crystal structure of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5XC1_A Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol [Bacillus sp. NG-27],5XC1_B Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol [Bacillus sp. NG-27] |
| 5EB8_A | 2.73e-17 | 451 | 747 | 85 | 350 | Crystalstructure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5EB8_B Crystal structure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O94163 | 3.87e-14 | 491 | 754 | 125 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| Q96VB6 | 6.66e-14 | 491 | 754 | 123 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| P07528 | 2.27e-13 | 451 | 747 | 130 | 391 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P14768 | 2.77e-13 | 511 | 757 | 381 | 611 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| O59859 | 9.72e-13 | 491 | 754 | 127 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
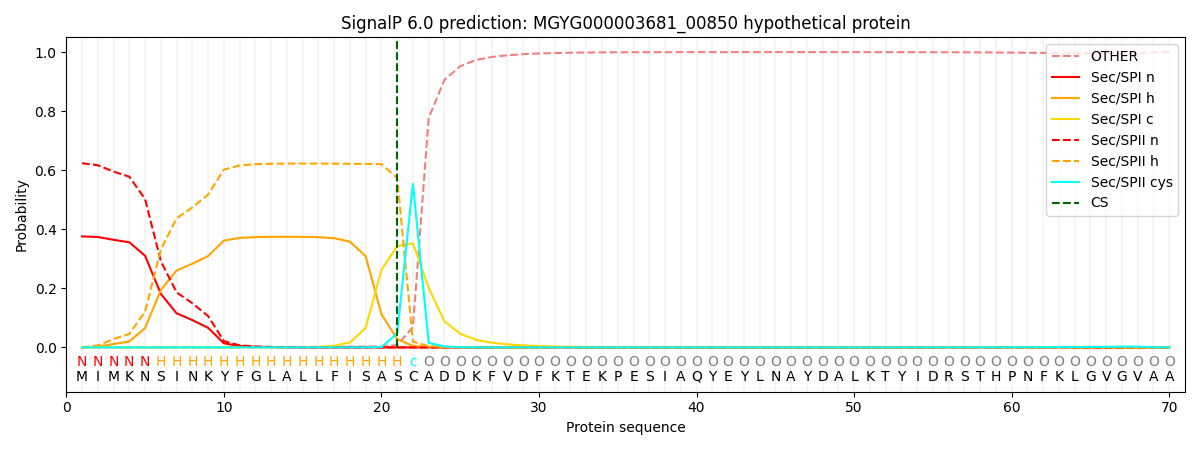
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000740 | 0.367136 | 0.631499 | 0.000199 | 0.000207 | 0.000201 |
