You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003681_01237
You are here: Home > Sequence: MGYG000003681_01237
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides stercoris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides stercoris | |||||||||||
| CAZyme ID | MGYG000003681_01237 | |||||||||||
| CAZy Family | GH125 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 77763; End: 79214 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH125 | 69 | 469 | 3e-181 | 0.9975124378109452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3538 | COG3538 | 0.0 | 47 | 479 | 1 | 430 | Meiotically up-regulated gene 157 (Mug157) protein (function unknown) [Function unknown]. |
| pfam06824 | Glyco_hydro_125 | 0.0 | 69 | 469 | 1 | 416 | Metal-independent alpha-mannosidase (GH125). This family, which contains bacterial and fungal glycoside hydrolases, is also known as GH125. They function as metal-independent alpha-mannosidases, with specificity for alpha-1,6-linked non-reducing terminal mannose residues. Structurally this family is part of the 6 hairpin glycosidase superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CUA18275.1 | 0.0 | 1 | 483 | 1 | 483 |
| QRP90519.1 | 0.0 | 1 | 483 | 1 | 483 |
| AKA51568.1 | 0.0 | 1 | 483 | 1 | 483 |
| QMI79243.1 | 4.16e-315 | 1 | 483 | 1 | 483 |
| QBJ17751.1 | 4.16e-315 | 1 | 483 | 1 | 483 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ON6_A | 1.98e-189 | 43 | 473 | 23 | 450 | Crystalstructure of an exo-alpha-1,6-mannosidase (bacova_03626) from bacteroides ovatus at 1.70 a resolution [Bacteroides ovatus ATCC 8483],3ON6_B Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03626) from bacteroides ovatus at 1.70 a resolution [Bacteroides ovatus ATCC 8483] |
| 2P0V_A | 3.73e-189 | 43 | 473 | 43 | 470 | Crystalstructure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 [Bacteroides thetaiotaomicron VPI-5482],2P0V_B Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 [Bacteroides thetaiotaomicron VPI-5482] |
| 3P2C_A | 4.28e-189 | 43 | 473 | 21 | 452 | Crystalstructure of an exo-alpha-1,6-mannosidase (bacova_03347) from bacteroides ovatus at 1.60 a resolution [Bacteroides ovatus ATCC 8483],3P2C_B Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03347) from bacteroides ovatus at 1.60 a resolution [Bacteroides ovatus ATCC 8483] |
| 3QT3_A | 1.43e-135 | 56 | 472 | 12 | 423 | Analysisof a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure [Clostridium perfringens],3QT9_A Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Clostridium perfringens CPE0426 complexed with alpha-1,6-linked 1-thio-alpha-mannobiose [Clostridium perfringens] |
| 6RQK_A | 1.86e-135 | 56 | 472 | 12 | 423 | Crystalstructure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole [Clostridium perfringens str. 13],6RQK_B Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole [Clostridium perfringens str. 13] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q10449 | 5.45e-123 | 43 | 479 | 51 | 504 | Meiotically up-regulated gene 157 protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mug157 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
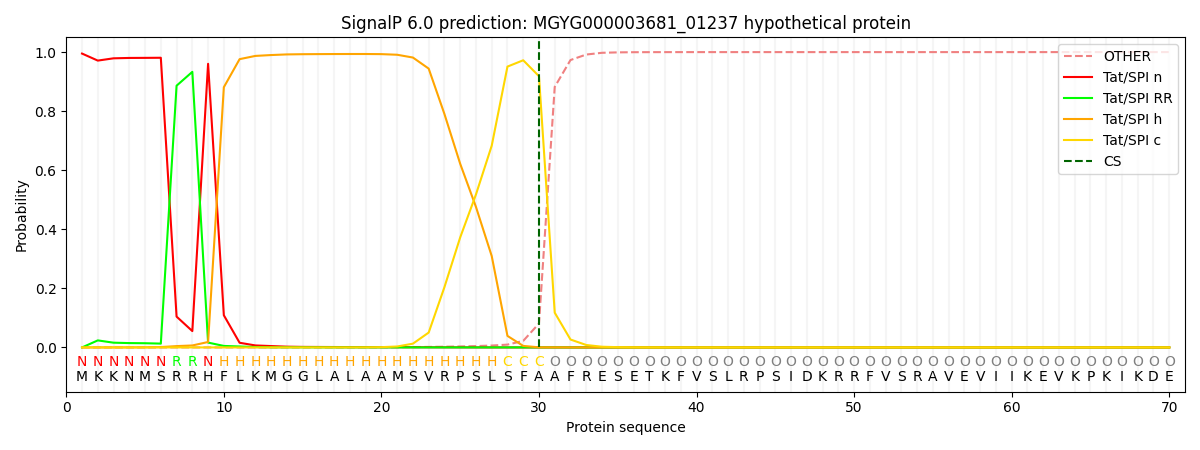
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000005 | 0.000010 | 0.000003 | 0.995047 | 0.004924 | 0.000000 |
