You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003687_00179
Basic Information
help
| Species |
Paenibacillus polymyxa
|
| Lineage |
Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus polymyxa
|
| CAZyme ID |
MGYG000003687_00179
|
| CAZy Family |
CBM22 |
| CAZyme Description |
Endo-1,4-beta-xylanase C
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003687 |
5656034 |
Isolate |
China |
Asia |
|
| Gene Location |
Start: 166273;
End: 166725
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| CBM22 |
52 |
129 |
1.4e-17 |
0.5801526717557252 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam02018
|
CBM_4_9 |
2.05e-15 |
56 |
136 |
7 |
86 |
Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
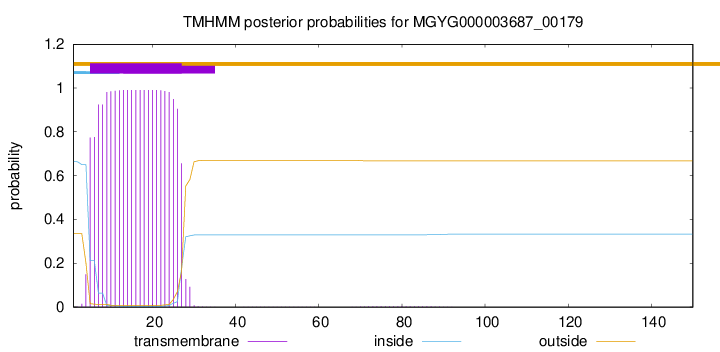
This protein is predicted as SP

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.174093
|
0.783006
|
0.040746
|
0.001068
|
0.000476
|
0.000578
|