You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003687_00362
You are here: Home > Sequence: MGYG000003687_00362
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus polymyxa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus polymyxa | |||||||||||
| CAZyme ID | MGYG000003687_00362 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 341245; End: 342201 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 52 | 314 | 9.8e-80 | 0.8910891089108911 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 1.63e-110 | 5 | 318 | 1 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 2.32e-95 | 33 | 313 | 16 | 309 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.17e-91 | 65 | 312 | 2 | 263 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIY09368.1 | 1.11e-240 | 1 | 318 | 1 | 318 |
| AZH31856.1 | 1.11e-240 | 1 | 318 | 1 | 318 |
| ADO59301.1 | 1.11e-240 | 1 | 318 | 1 | 318 |
| CCI71792.1 | 1.11e-240 | 1 | 318 | 1 | 318 |
| QPK52792.1 | 1.11e-240 | 1 | 318 | 1 | 318 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PMY_A | 5.94e-95 | 33 | 316 | 5 | 299 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMY_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMZ_A Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306],4PMZ_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306] |
| 4PN2_A | 6.34e-95 | 33 | 316 | 6 | 300 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306],4PN2_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306] |
| 4PMX_A | 6.55e-95 | 33 | 316 | 6 | 300 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form [Xanthomonas citri pv. citri str. 306] |
| 4XUY_A | 9.78e-35 | 33 | 308 | 12 | 294 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 1NQ6_A | 7.54e-33 | 55 | 308 | 37 | 294 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23030 | 9.24e-69 | 37 | 312 | 323 | 592 | Endo-1,4-beta-xylanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynB PE=1 SV=2 |
| O94163 | 3.23e-35 | 7 | 308 | 10 | 319 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| P33559 | 1.23e-34 | 33 | 308 | 37 | 319 | Endo-1,4-beta-xylanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynA PE=1 SV=2 |
| A2QFV7 | 9.11e-34 | 33 | 308 | 37 | 319 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
| C5J411 | 1.27e-33 | 33 | 308 | 37 | 319 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
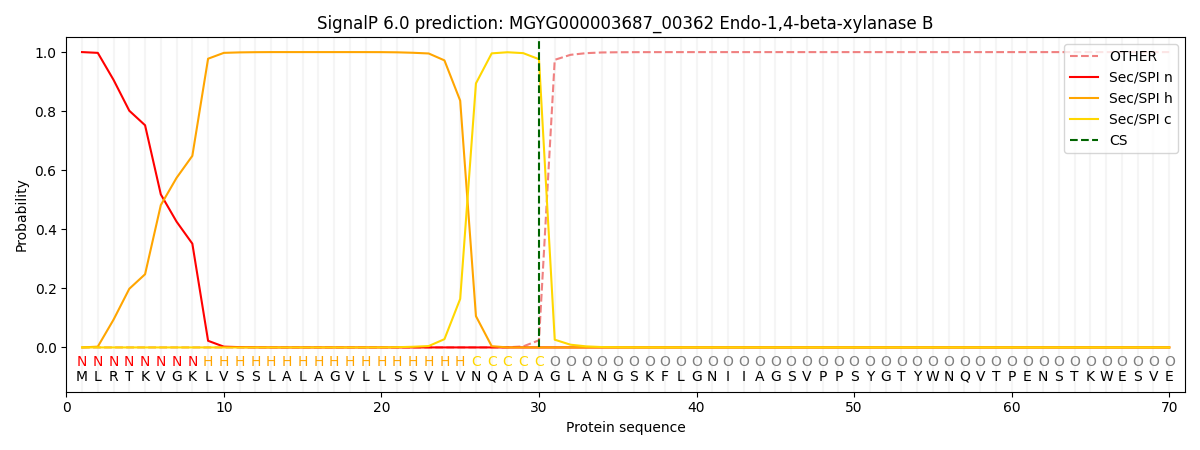
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999081 | 0.000172 | 0.000179 | 0.000162 | 0.000144 |
