You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003687_01201
You are here: Home > Sequence: MGYG000003687_01201
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
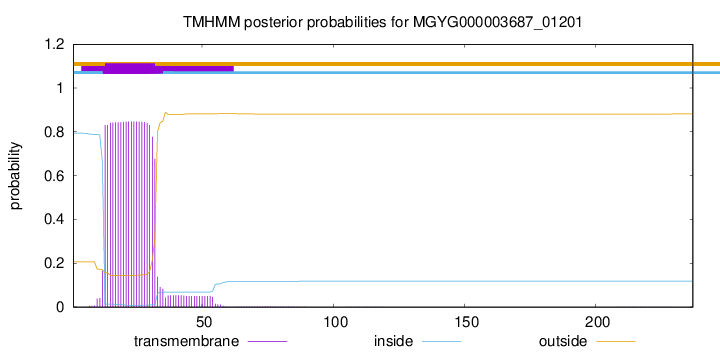
TMHMM annotations
Basic Information help
| Species | Paenibacillus polymyxa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus polymyxa | |||||||||||
| CAZyme ID | MGYG000003687_01201 | |||||||||||
| CAZy Family | CBM63 | |||||||||||
| CAZyme Description | Expansin-YoaJ | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 280855; End: 281568 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM63 | 144 | 219 | 8.2e-39 | 0.9743589743589743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4305 | YoaJ | 2.37e-132 | 18 | 237 | 13 | 232 | Peptidoglycan-binding domain, expansin [Cell wall/membrane/envelope biogenesis]. |
| cd22272 | DPBB_EXLX1-like | 2.56e-54 | 34 | 134 | 1 | 101 | N-terminal double-psi beta-barrel fold domain of bacterial expansins similar to Bacillus subtilis EXLX1. This subfamily is composed of bacterial expansins including Bacillus subtilis EXLX1, also called expansin-YoaJ. Similar to plant expansins, EXLX1 contains an N-terminal domain (D1) homologous to the catalytic domain of glycoside hydrolase family 45 (GH45) proteins but with no hydrolytic activity, and a C-terminal domain (D2) homologous to group-2 grass pollen allergens. It strongly binds to crystalline cellulose via D2, and weakly binds soluble cellooligosaccharides. Bacterial expansins, which are present in some plant pathogens, have the ability to loosen plant cell walls, but with weaker activity compared to plant expansins. They may have a role in plant-bacterial interactions. This model represents the N-terminal domain of EXLX1 and similar bacterial expansins, which adopts a double-psi beta-barrel (DPBB) fold. |
| cd22191 | DPBB_RlpA_EXP_N-like | 1.31e-17 | 36 | 134 | 1 | 94 | double-psi beta-barrel fold of RlpA, N-terminal domain of expansins, and similar domains. The double-psi beta-barrel (DPBB) fold is found in a divergent group of proteins, including endolytic peptidoglycan transglycosylase RlpA (rare lipoprotein A), EG45-like domain containing proteins, kiwellins, Streptomyces papain inhibitor (SPI), and the N-terminal domain of plant and bacterial expansins. RlpA may work in tandem with amidases to degrade peptidoglycan (PG) in the division septum and lateral wall to facilitate daughter cell separation. An EG45-like domain containing protein from Arabidopsis thaliana, called plant natriuretic peptide A (AtPNP-A), functions in cell volume regulation. Kiwellin proteins comprise a widespread family of plant-defense proteins that target pathogenic bacterial/fungal effectors that down-regulate plant defense responses. SPI is a stress protein produced under hyperthermal stress conditions that serves as a glutamine and lysine donor substrate for microbial transglutaminase (MTG, EC 2.3.2.13) from Streptomycetes. Some expansin family proteins display cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. |
| cd22271 | DPBB_EXP_N-like | 3.39e-11 | 26 | 135 | 1 | 115 | N-terminal double-psi beta-barrel fold domain of the expansin family and similar domains. The plant expansin family consists of four subfamilies, alpha-expansin (EXPA), beta-expansin (EXPB), expansin-like A (EXLA), and expansin-like B (EXLB). EXPA and EXPB display cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. EXPA proteins function more efficiently on dicotyledonous cell walls, whereas EXPB proteins exhibit specificity for the cell walls of monocotyledons. Expansins also affect environmental stress responses. Expansin family proteins contain an N-terminal domain (D1) homologous to the catalytic domain of glycoside hydrolase family 45 (GH45) proteins but with no hydrolytic activity, and a C-terminal domain (D2) homologous to group-2 grass pollen allergens. This family also includes GH45 endoglucanases from mollusks. This model represents the N-terminal domain of expansins and similar proteins, which adopts a double-psi beta-barrel (DPBB) fold. |
| PLN03024 | PLN03024 | 2.08e-05 | 1 | 132 | 1 | 123 | Putative EG45-like domain containing protein 1; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDA28429.1 | 2.82e-173 | 1 | 237 | 1 | 237 |
| QPK59247.1 | 2.82e-173 | 1 | 237 | 1 | 237 |
| QPK54157.1 | 2.82e-173 | 1 | 237 | 1 | 237 |
| AUS27696.1 | 8.08e-173 | 1 | 237 | 1 | 237 |
| QOH63021.1 | 1.15e-172 | 1 | 237 | 1 | 237 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3D30_A | 3.43e-116 | 31 | 237 | 2 | 208 | Structureof an expansin like protein from Bacillus Subtilis at 1.9A resolution [Bacillus subtilis],4FER_A Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose [Bacillus subtilis subsp. subtilis str. 168],4FER_B Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose [Bacillus subtilis subsp. subtilis str. 168],4FFT_A Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with mixed-linkage glucan [Bacillus subtilis subsp. subtilis str. 168],4FFT_B Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with mixed-linkage glucan [Bacillus subtilis subsp. subtilis str. 168],4FG2_A Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellotetraose [Bacillus subtilis subsp. subtilis str. 168],4FG2_B Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellotetraose [Bacillus subtilis subsp. subtilis str. 168],4FG4_A Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with hemithiocellodextrin [Bacillus subtilis subsp. subtilis str. 168],4FG4_B Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with hemithiocellodextrin [Bacillus subtilis subsp. subtilis str. 168] |
| 2BH0_A | 9.37e-114 | 31 | 237 | 2 | 208 | Crystalstructure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom [Bacillus subtilis] |
| 4JCW_A | 4.45e-26 | 37 | 208 | 8 | 176 | Crystalstructure of Clavibacter michiganensis expansin in complex with cellopentaose [Clavibacter michiganensis subsp. michiganensis NCPPB 382],4JCW_B Crystal structure of Clavibacter michiganensis expansin in complex with cellopentaose [Clavibacter michiganensis subsp. michiganensis NCPPB 382],4JJO_A crystal structure of apo-clavibacter Michiganensis expansin [Clavibacter michiganensis] |
| 4JS7_A | 2.42e-25 | 37 | 208 | 8 | 176 | Crystalstructure of D78N mutant apo form of clavibacter michiganensis expansin [Clavibacter michiganensis subsp. michiganensis NCPPB 382],4L48_A Crystal structure of d78n mutant clavibacter michiganensis expansin in complex with cellohexaose [Clavibacter michiganensis subsp. michiganensis NCPPB 382],4L48_C Crystal structure of d78n mutant clavibacter michiganensis expansin in complex with cellohexaose [Clavibacter michiganensis subsp. michiganensis NCPPB 382] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34918 | 6.81e-119 | 18 | 237 | 13 | 232 | Expansin-YoaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yoaJ PE=1 SV=1 |
| Q55G31 | 1.23e-11 | 39 | 236 | 38 | 244 | Expansin-like protein 1 OS=Dictyostelium discoideum OX=44689 GN=expl1 PE=2 SV=1 |
| Q7KWS2 | 1.42e-10 | 9 | 216 | 2 | 218 | Expansin-like protein 3 OS=Dictyostelium discoideum OX=44689 GN=expl3 PE=2 SV=1 |
| Q86AV4 | 1.18e-09 | 58 | 221 | 59 | 225 | Expansin-like protein 5 OS=Dictyostelium discoideum OX=44689 GN=expl5 PE=2 SV=1 |
| Q55G32 | 4.59e-06 | 77 | 233 | 76 | 247 | Expansin-like protein 6 OS=Dictyostelium discoideum OX=44689 GN=expl6 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
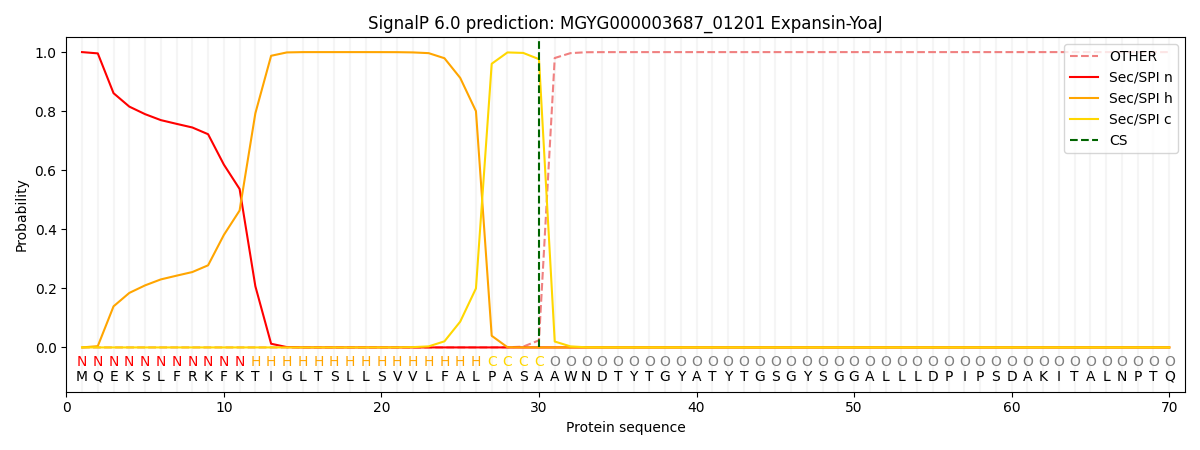
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000364 | 0.998907 | 0.000222 | 0.000168 | 0.000169 | 0.000160 |