You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003690_01000
You are here: Home > Sequence: MGYG000003690_01000
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
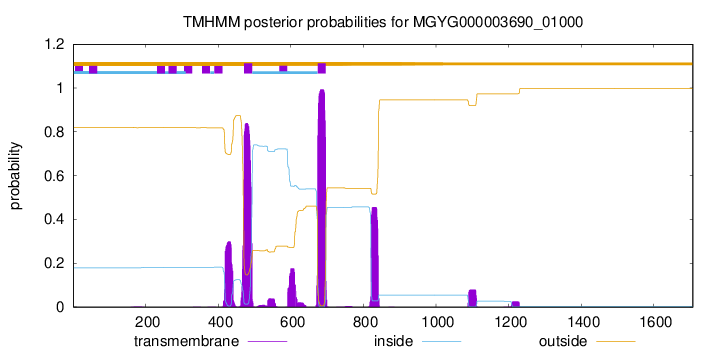
TMHMM annotations
Basic Information help
| Species | Lactobacillus gasseri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus gasseri | |||||||||||
| CAZyme ID | MGYG000003690_01000 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57155; End: 62281 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0791 | Spr | 2.09e-19 | 1272 | 1422 | 39 | 191 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| pfam00877 | NLPC_P60 | 3.73e-19 | 1320 | 1398 | 1 | 82 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 1.33e-16 | 1283 | 1397 | 342 | 457 | invasion associated endopeptidase. |
| COG5283 | COG5283 | 4.46e-11 | 15 | 1287 | 4 | 1160 | Phage-related tail protein [Mobilome: prophages, transposons]. |
| NF033742 | NlpC_p60_RipB | 1.50e-09 | 1320 | 1398 | 89 | 181 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHJ74970.1 | 0.0 | 1 | 1708 | 1 | 1694 |
| QTP20680.1 | 0.0 | 1 | 1708 | 1 | 1672 |
| ART99103.1 | 0.0 | 1 | 1708 | 1 | 1672 |
| AZZ67568.1 | 0.0 | 1 | 1708 | 1 | 1759 |
| QTP20241.1 | 6.46e-190 | 157 | 1597 | 266 | 1722 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CFL_A | 4.70e-11 | 1308 | 1401 | 14 | 114 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4XCM_A | 7.70e-08 | 1308 | 1398 | 115 | 205 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 3PBI_A | 8.17e-06 | 1320 | 1415 | 97 | 201 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01835 | 5.67e-11 | 1305 | 1402 | 394 | 492 | Probable endopeptidase p60 OS=Listeria grayi OX=1641 GN=iap PE=3 SV=1 |
| Q8NNK6 | 3.19e-08 | 1305 | 1389 | 95 | 179 | Probable endopeptidase Cgl2188 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=Cgl2188 PE=3 SV=1 |
| A4QFQ3 | 3.30e-08 | 1305 | 1389 | 97 | 181 | Probable endopeptidase cgR_2070 OS=Corynebacterium glutamicum (strain R) OX=340322 GN=cgR_2070 PE=1 SV=1 |
| P96740 | 1.77e-07 | 1306 | 1396 | 165 | 260 | Gamma-DL-glutamyl hydrolase OS=Bacillus subtilis (strain 168) OX=224308 GN=pgdS PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
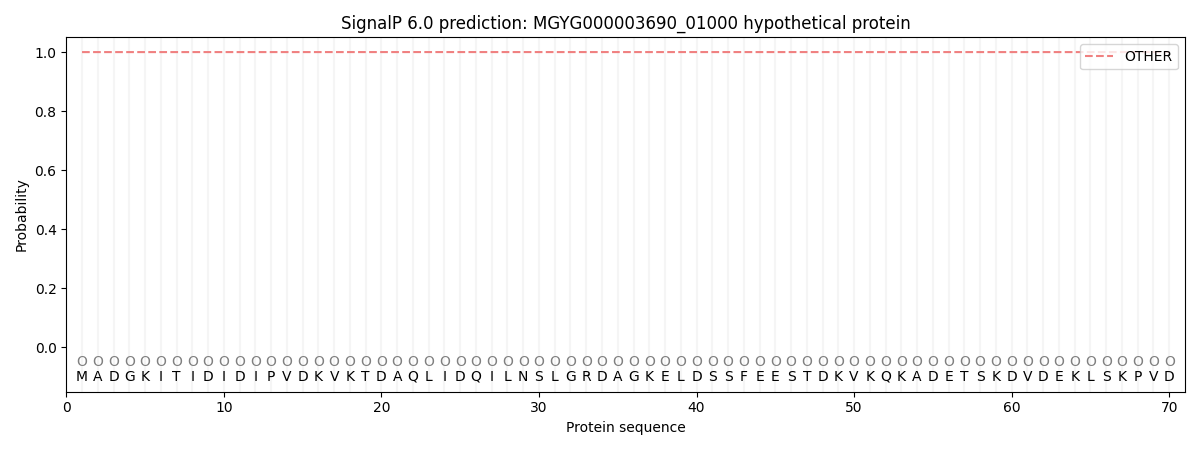
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000060 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |