You are browsing environment: HUMAN GUT
MGYG000003701_02771
Basic Information
help
Species
Parabacteroides sp003480915
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp003480915
CAZyme ID
MGYG000003701_02771
CAZy Family
CBM51
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003701
6084075
Isolate
China
Asia
Gene Location
Start: 92751;
End: 95582
Strand: +
No EC number prediction in MGYG000003701_02771.
Family
Start
End
Evalue
family coverage
CBM51
130
201
8.3e-18
0.5298507462686567
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam08305
NPCBM
2.03e-35
29
202
5
136
NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.)
more
smart00776
NPCBM
1.01e-26
29
202
7
145
This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
7JS4_A
9.59e-07
125
318
676
859
ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124]
more
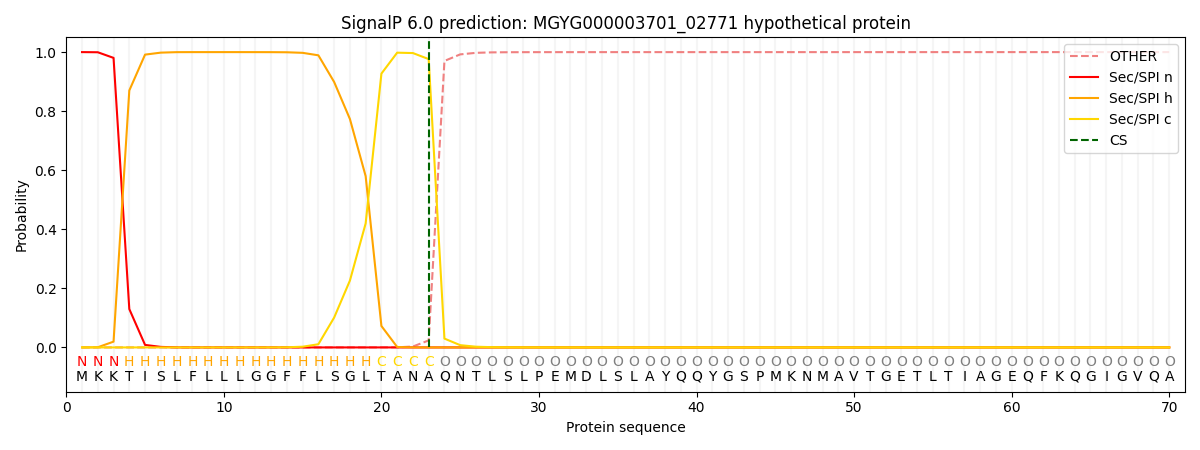
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000277
0.998941
0.000243
0.000181
0.000165
0.000142