You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003711_00208
Basic Information
help
| Species |
Stenotrophomonas maltophilia_AQ
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_AQ
|
| CAZyme ID |
MGYG000003711_00208
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 611 |
|
66751.89 |
9.7047 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003711 |
4541486 |
MAG |
Estonia |
Europe |
|
| Gene Location |
Start: 8581;
End: 10416
Strand: +
|
No EC number prediction in MGYG000003711_00208.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
7 |
370 |
3.7e-25 |
0.6666666666666666 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
9.11e-27 |
11 |
350 |
19 |
372 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
7.27e-17 |
51 |
211 |
1 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| pfam17957
|
Big_7 |
9.66e-06 |
505 |
587 |
1 |
63 |
Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
This protein is predicted as OTHER
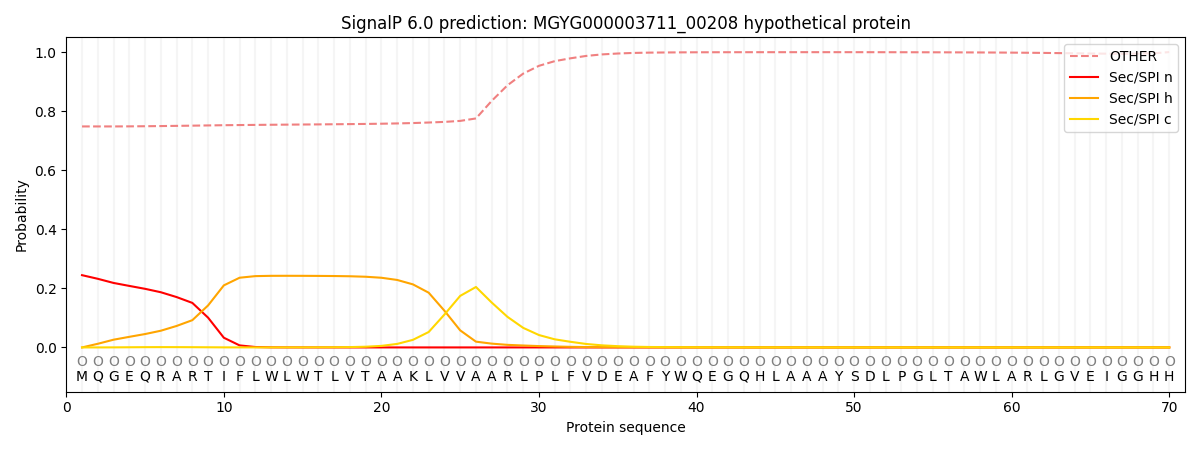
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.762298
|
0.225995
|
0.002734
|
0.000879
|
0.000634
|
0.007464
|
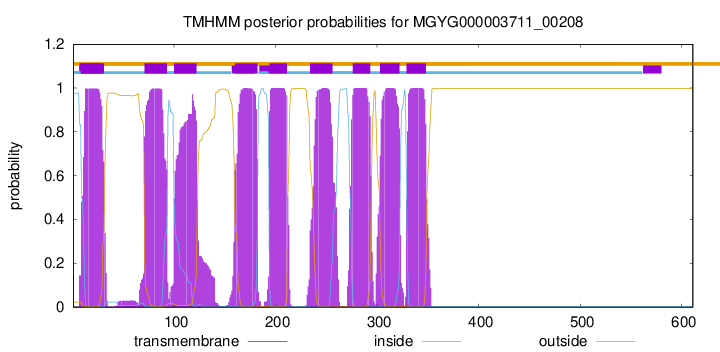
| start |
end |
| 9 |
31 |
| 71 |
93 |
| 100 |
122 |
| 160 |
182 |
| 194 |
211 |
| 234 |
256 |
| 276 |
293 |
| 303 |
322 |
| 329 |
348 |


