You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003712_00127
You are here: Home > Sequence: MGYG000003712_00127
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Serratia liquefaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Serratia; Serratia liquefaciens | |||||||||||
| CAZyme ID | MGYG000003712_00127 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Chitinase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12886; End: 14577 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 158 | 547 | 9.7e-72 | 0.9459459459459459 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 9.91e-134 | 160 | 544 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| smart00636 | Glyco_18 | 1.31e-128 | 158 | 544 | 1 | 334 | Glyco_18 domain. |
| COG3325 | ChiA | 6.92e-123 | 127 | 557 | 7 | 437 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| pfam00704 | Glyco_hydro_18 | 9.27e-96 | 158 | 544 | 1 | 307 | Glycosyl hydrolases family 18. |
| pfam08329 | ChitinaseA_N | 2.40e-69 | 24 | 154 | 1 | 130 | Chitinase A, N-terminal domain. This domain is found in a number of bacterial chitinases and similar viral proteins. It is organized into a fibronectin III module domain-like fold, comprising only beta strands. Its function is not known, but it may be involved in interaction with the enzyme substrate, chitin. It is separated by a hinge region from the catalytic domain (pfam00704); this hinge region is probably mobile, allowing the N-terminal domain to have different relative positions in solution. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDL34465.1 | 0.0 | 1 | 563 | 1 | 563 |
| AGQ28968.1 | 0.0 | 1 | 563 | 1 | 563 |
| AMG99785.1 | 0.0 | 1 | 563 | 1 | 563 |
| AYO40583.1 | 0.0 | 1 | 563 | 1 | 563 |
| QNQ54526.1 | 0.0 | 1 | 563 | 1 | 563 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WLY_A | 0.0 | 24 | 561 | 1 | 538 | ChitinaseA from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. [Serratia marcescens],2WLZ_A Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. [Serratia marcescens],2WM0_A Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline thioamide. [Serratia marcescens] |
| 1NH6_A | 0.0 | 24 | 561 | 1 | 538 | ChainA, chitinase A [Serratia marcescens] |
| 1RD6_A | 0.0 | 1 | 561 | 1 | 561 | ChainA, Chitinase A [Serratia marcescens],1X6L_A Chain A, Chitinase A [Serratia marcescens],1X6N_A Chain A, Chitinase A [Serratia marcescens] |
| 2WK2_A | 0.0 | 24 | 561 | 1 | 538 | ChitinaseA from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. [Serratia marcescens] |
| 1EDQ_A | 0.0 | 24 | 561 | 1 | 538 | CrystalStructure Of Chitinase A From S. Marcescens At 1.55 Angstroms [Serratia marcescens],1FFQ_A Crystal Structure Of Chitinase A Complexed With Allosamidin [Serratia marcescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07254 | 0.0 | 1 | 561 | 1 | 561 | Chitinase A OS=Serratia marcescens OX=615 GN=chiA PE=1 SV=3 |
| P41684 | 2.38e-240 | 20 | 554 | 13 | 543 | Chitinase OS=Autographa californica nuclear polyhedrosis virus OX=46015 GN=CHIA PE=1 SV=1 |
| O10363 | 5.94e-231 | 13 | 553 | 5 | 541 | Probable endochitinase OS=Orgyia pseudotsugata multicapsid polyhedrosis virus OX=262177 GN=ORF124 PE=3 SV=1 |
| P32823 | 2.80e-181 | 3 | 556 | 2 | 585 | Chitinase A OS=Pseudoalteromonas piscicida OX=43662 GN=chiA PE=1 SV=1 |
| P20533 | 1.48e-54 | 155 | 551 | 42 | 445 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
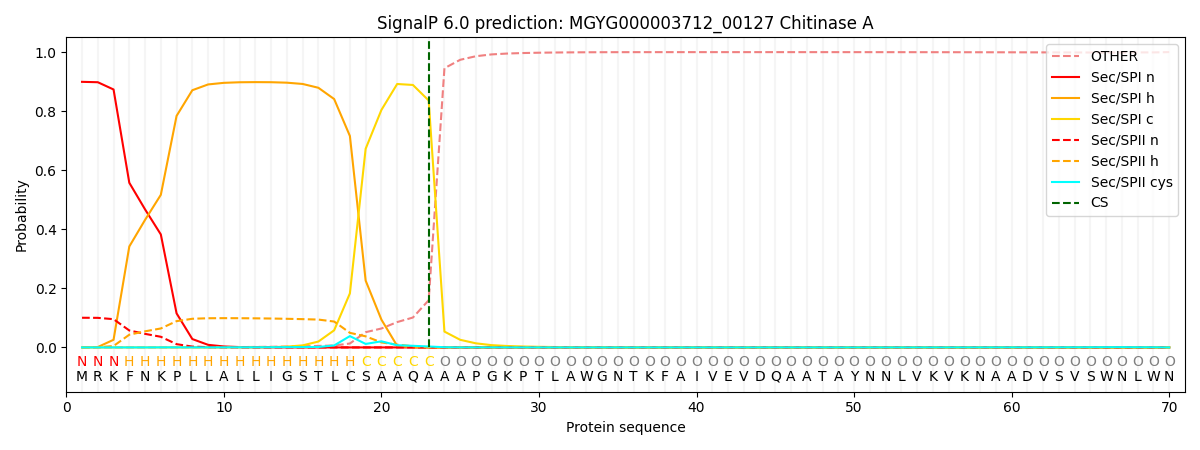
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001032 | 0.892306 | 0.105472 | 0.000570 | 0.000305 | 0.000276 |
