You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003713_00807
You are here: Home > Sequence: MGYG000003713_00807
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
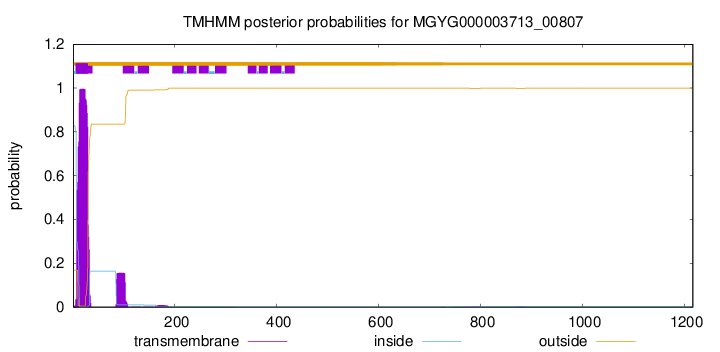
TMHMM annotations
Basic Information help
| Species | Campylobacter_B sp900539505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Campylobacteraceae; Campylobacter_B; Campylobacter_B sp900539505 | |||||||||||
| CAZyme ID | MGYG000003713_00807 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24000; End: 27653 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 59 | 225 | 9.7e-45 | 0.8813559322033898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4953 | PbpC | 6.68e-75 | 8 | 272 | 3 | 261 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| TIGR02073 | PBP_1c | 1.26e-71 | 618 | 966 | 242 | 593 | penicillin-binding protein 1C. This subfamily of the penicillin binding proteins includes the member from E. coli designated penicillin-binding protein 1C. Members have both transglycosylase and transpeptidase domains and are involved in forming cross-links in the late stages of peptidoglycan biosynthesis. All members of this subfamily are presumed to have the same basic function. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG4953 | PbpC | 2.85e-65 | 612 | 959 | 258 | 602 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| COG0744 | MrcB | 1.92e-52 | 1 | 276 | 14 | 282 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG0744 | MrcB | 2.84e-51 | 556 | 909 | 228 | 592 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKT92627.1 | 0.0 | 1 | 1217 | 1 | 1185 |
| QKF71374.1 | 7.52e-213 | 5 | 1050 | 6 | 694 |
| ABS52052.1 | 2.39e-194 | 5 | 1036 | 2 | 673 |
| QKF86135.1 | 6.71e-190 | 5 | 1050 | 2 | 694 |
| QCD46888.1 | 4.79e-163 | 2 | 1034 | 7 | 677 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 4.72e-22 | 62 | 246 | 20 | 198 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 6.42e-22 | 62 | 246 | 20 | 198 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3UDF_A | 5.83e-20 | 69 | 251 | 46 | 222 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 4OON_A | 2.68e-17 | 69 | 251 | 46 | 222 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3DWK_A | 5.55e-16 | 43 | 210 | 10 | 168 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P76577 | 2.36e-35 | 25 | 291 | 29 | 286 | Penicillin-binding protein 1C OS=Escherichia coli (strain K12) OX=83333 GN=pbpC PE=1 SV=1 |
| P40750 | 5.34e-22 | 24 | 248 | 37 | 256 | Penicillin-binding protein 4 OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpD PE=1 SV=2 |
| A7FY32 | 2.39e-20 | 15 | 262 | 30 | 279 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 2.39e-20 | 15 | 262 | 30 | 279 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 3.14e-20 | 15 | 262 | 30 | 279 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
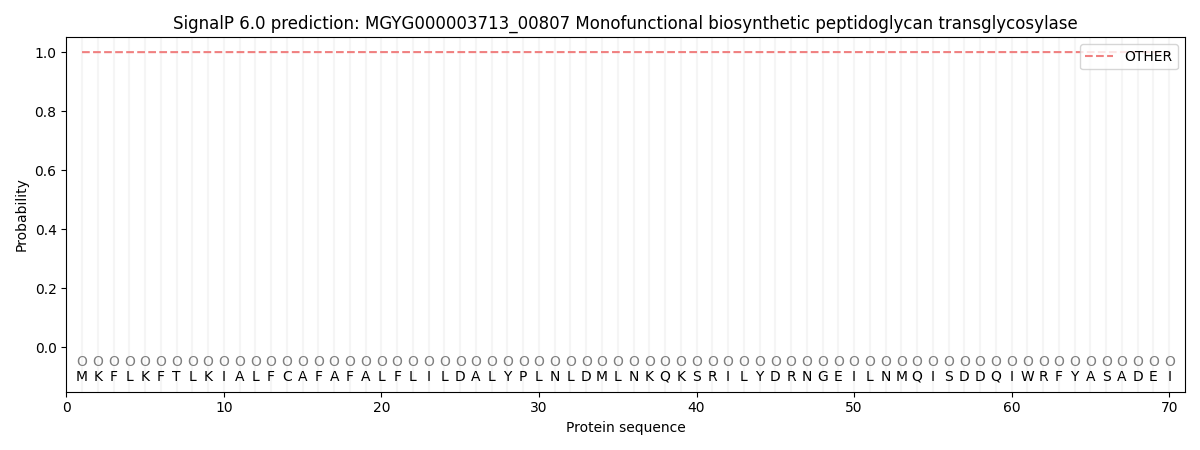
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999854 | 0.000101 | 0.000020 | 0.000000 | 0.000000 | 0.000060 |