You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003723_00366
You are here: Home > Sequence: MGYG000003723_00366
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Mobiluncus; | |||||||||||
| CAZyme ID | MGYG000003723_00366 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42884; End: 44131 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04122 | CW_binding_2 | 5.12e-16 | 197 | 277 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 7.72e-14 | 99 | 182 | 2 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| COG2247 | LytB | 1.51e-11 | 107 | 217 | 16 | 125 | Putative cell wall-binding domain [Cell wall/membrane/envelope biogenesis]. |
| NF033435 | S-layer_Clost | 3.18e-06 | 105 | 243 | 404 | 553 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADI66456.1 | 5.40e-243 | 1 | 415 | 1 | 419 |
| QQU08026.1 | 5.40e-243 | 1 | 415 | 1 | 419 |
| QQT13535.1 | 2.20e-242 | 1 | 415 | 1 | 419 |
| QEW01478.1 | 4.20e-37 | 318 | 415 | 106 | 204 |
| QIG41297.1 | 7.33e-37 | 318 | 415 | 102 | 200 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J72_A | 2.31e-14 | 99 | 265 | 135 | 308 | ChainA, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630],5J72_B Chain B, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630] |
| 5J6Q_A | 1.88e-09 | 99 | 274 | 289 | 473 | ChainA, Cell wall binding protein cwp8 [Clostridioides difficile 630] |
| 7ACZ_B | 4.39e-09 | 121 | 239 | 76 | 205 | ChainB, SLPH (HMW SLP) [Clostridioides difficile R20291],7ACZ_D Chain D, SLPH (HMW SLP) [Clostridioides difficile R20291] |
| 7ACX_B | 1.22e-08 | 105 | 241 | 60 | 207 | ChainB, S-layer protein [Clostridioides difficile],7ACX_D Chain D, S-layer protein [Clostridioides difficile] |
| 7ACY_B | 1.22e-08 | 105 | 241 | 60 | 207 | ChainB, S-layer protein [Clostridioides difficile 630],7ACY_D Chain D, S-layer protein [Clostridioides difficile 630],7QGQ_B Chain B, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_D Chain D, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_J Chain J, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_K Chain K, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_L Chain L, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_M Chain M, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_N Chain N, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_T Chain T, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_U Chain U, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_V Chain V, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_W Chain W, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_X Chain X, Precursor of the S-layer proteins [Clostridioides difficile 630] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02114 | 2.40e-14 | 99 | 276 | 31 | 212 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| Q02113 | 2.17e-10 | 101 | 277 | 67 | 239 | Amidase enhancer OS=Bacillus subtilis (strain 168) OX=224308 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
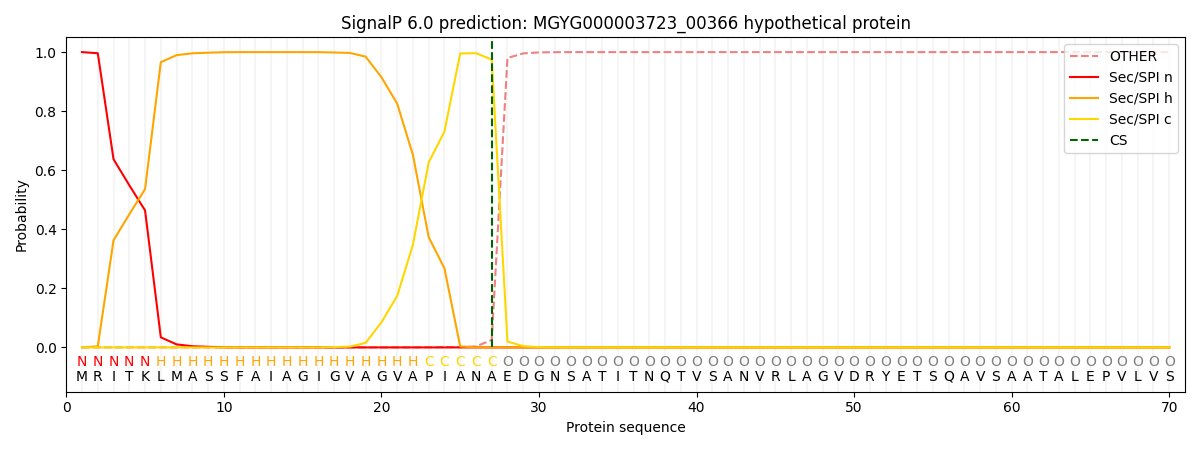
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000315 | 0.998861 | 0.000214 | 0.000215 | 0.000209 | 0.000171 |
