You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003733_01275
You are here: Home > Sequence: MGYG000003733_01275
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp003020075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp003020075 | |||||||||||
| CAZyme ID | MGYG000003733_01275 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17351; End: 18616 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 73 | 387 | 2.8e-50 | 0.9590443686006825 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.72e-15 | 75 | 256 | 31 | 203 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUC94471.1 | 9.81e-254 | 1 | 414 | 1 | 414 |
| AOO81558.1 | 1.98e-195 | 3 | 416 | 295 | 704 |
| AZO80994.1 | 7.06e-195 | 1 | 416 | 1 | 414 |
| BCB21207.1 | 1.92e-185 | 1 | 414 | 1 | 405 |
| ACB78436.1 | 2.36e-96 | 3 | 414 | 7 | 415 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KDD_A | 4.28e-07 | 77 | 398 | 50 | 304 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 1CEC_A | 3.41e-06 | 77 | 261 | 37 | 213 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 8.01e-06 | 77 | 261 | 37 | 213 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as TAT
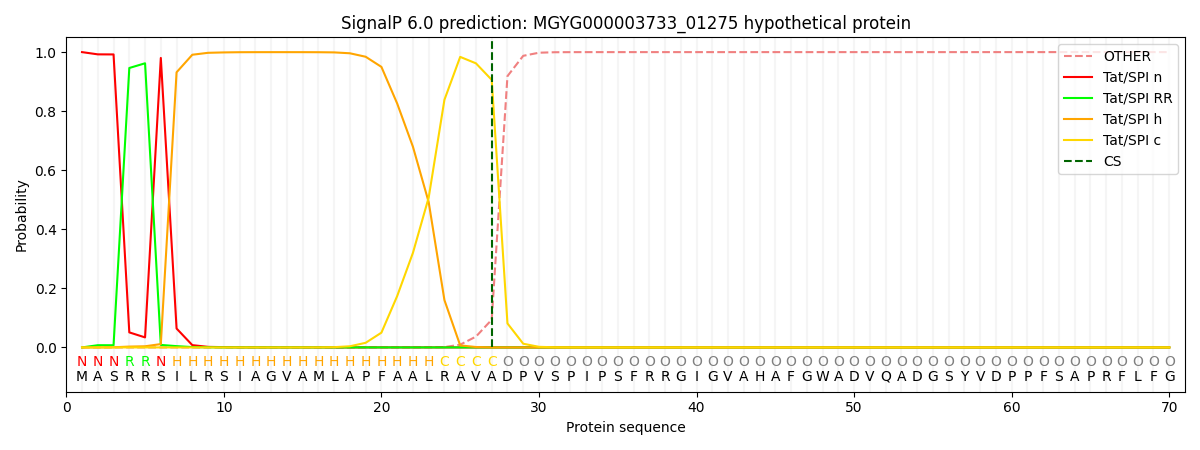
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999974 | 0.000000 | 0.000000 |
