You are browsing environment: HUMAN GUT
MGYG000003733_03373
Basic Information
help
Species
Bradyrhizobium sp003020075
Lineage
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp003020075
CAZyme ID
MGYG000003733_03373
CAZy Family
GT22
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
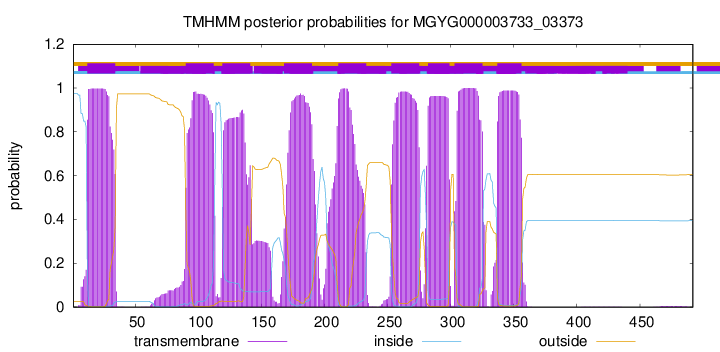
CGC
Molecular Weight
Isoelectric Point
492
53005.62
9.2232
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003733
7397431
MAG
Canada
North America
Gene Location
Start: 10462;
End: 11940
Strand: +
No EC number prediction in MGYG000003733_03373.
Family
Start
End
Evalue
family coverage
GT22
16
355
1.3e-49
0.8688946015424165
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam03901
Glyco_transf_22
2.11e-11
28
353
15
360
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
PLN02816
PLN02816
0.003
35
73
62
100
mannosyltransferase
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
Q6CAB8
2.83e-12
13
346
16
357
GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1
more
Q1LZA0
4.84e-06
35
342
74
382
GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1
more
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.001157
0.998012
0.000206
0.000202
0.000200
0.000202
start
end
12
34
90
112
119
141
168
190
211
233
253
275
282
299
303
325
337
356