You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003733_06647
You are here: Home > Sequence: MGYG000003733_06647
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp003020075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp003020075 | |||||||||||
| CAZyme ID | MGYG000003733_06647 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1979; End: 3541 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 49 | 294 | 2e-25 | 0.9485530546623794 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5309 | Scw11 | 1.19e-49 | 52 | 287 | 63 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| pfam00332 | Glyco_hydro_17 | 7.07e-08 | 125 | 293 | 106 | 305 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOZ20668.1 | 0.0 | 1 | 520 | 1 | 520 |
| BBB96363.1 | 0.0 | 1 | 520 | 1 | 520 |
| QOZ38433.1 | 0.0 | 1 | 520 | 1 | 523 |
| QIG99601.1 | 0.0 | 1 | 520 | 1 | 520 |
| AUC93157.1 | 0.0 | 1 | 520 | 1 | 523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FCG_A | 1.91e-06 | 25 | 290 | 46 | 379 | Crystalstructure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_B Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_C Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_D Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_E Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_F Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P43070 | 3.03e-18 | 57 | 293 | 44 | 301 | Glucan 1,3-beta-glucosidase OS=Candida albicans OX=5476 GN=BGL2 PE=3 SV=1 |
| Q5AMT2 | 1.01e-17 | 57 | 293 | 44 | 301 | Glucan 1,3-beta-glucosidase BGL2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BGL2 PE=1 SV=2 |
| P53334 | 6.05e-16 | 28 | 287 | 133 | 386 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
| P15703 | 5.65e-15 | 57 | 293 | 49 | 306 | Glucan 1,3-beta-glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=BGL2 PE=1 SV=1 |
| Q59XX2 | 1.43e-13 | 31 | 286 | 129 | 378 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
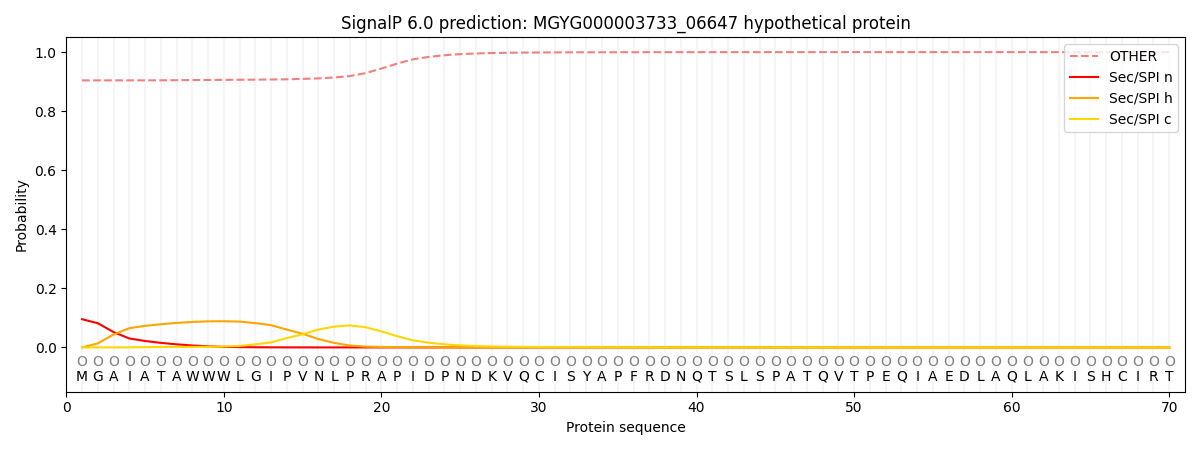
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.912519 | 0.086866 | 0.000266 | 0.000151 | 0.000077 | 0.000145 |
TMHMM Annotations download full data without filtering help
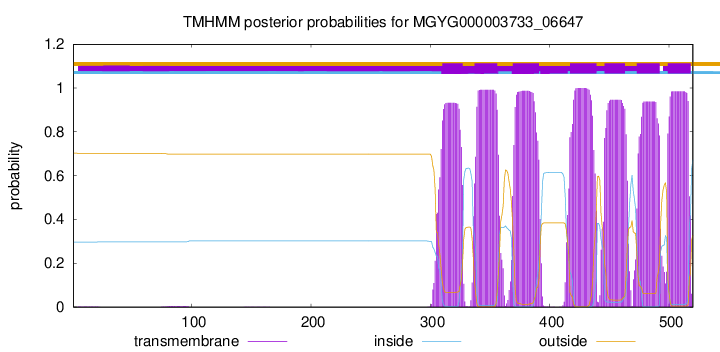
| start | end |
|---|---|
| 310 | 327 |
| 337 | 356 |
| 369 | 391 |
| 417 | 439 |
| 446 | 463 |
| 473 | 492 |
| 499 | 518 |
