You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003734_00067
You are here: Home > Sequence: MGYG000003734_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Afipia broomeae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Afipia; Afipia broomeae | |||||||||||
| CAZyme ID | MGYG000003734_00067 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73948; End: 75201 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 60 | 383 | 6.4e-67 | 0.9863481228668942 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.47e-08 | 66 | 387 | 27 | 272 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWN42027.1 | 2.39e-106 | 1 | 410 | 1 | 412 |
| QAZ42315.1 | 5.66e-67 | 18 | 411 | 16 | 414 |
| QGY02672.1 | 3.03e-66 | 28 | 410 | 31 | 410 |
| AZO00885.1 | 4.16e-66 | 30 | 410 | 26 | 412 |
| ACB78436.1 | 5.69e-66 | 7 | 414 | 15 | 419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NCO_A | 4.47e-14 | 23 | 409 | 8 | 319 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
| 6KDD_A | 1.12e-13 | 64 | 409 | 42 | 319 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 3RJX_A | 1.46e-13 | 23 | 409 | 8 | 319 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
| 3RJY_A | 2.64e-13 | 22 | 409 | 13 | 319 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
| 6UJE_A | 3.01e-10 | 164 | 408 | 124 | 308 | ChainA, Endoglucanase [Clostridioides difficile],6UJF_A Chain A, Endoglucanase [Clostridioides difficile] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14250 | 5.16e-11 | 98 | 386 | 377 | 636 | Endoglucanase 3 OS=Fibrobacter succinogenes (strain ATCC 19169 / S85) OX=59374 GN=cel-3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
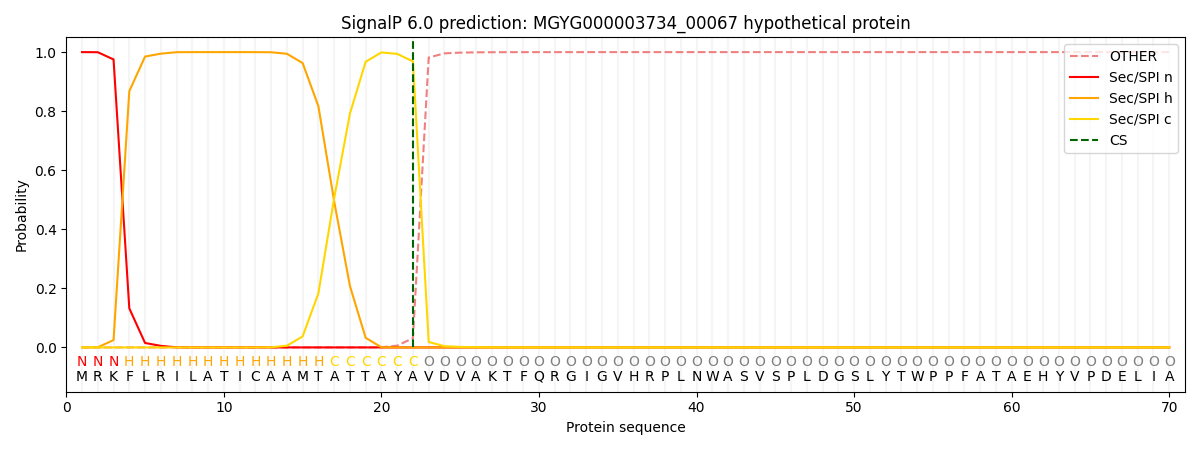
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000456 | 0.998660 | 0.000262 | 0.000212 | 0.000204 | 0.000172 |
