You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003742_00681
You are here: Home > Sequence: MGYG000003742_00681
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella buccalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella buccalis | |||||||||||
| CAZyme ID | MGYG000003742_00681 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15107; End: 16645 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 75 | 503 | 2.6e-136 | 0.988009592326139 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 2.30e-43 | 86 | 504 | 45 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02055 | Glyco_hydro_30 | 1.46e-32 | 88 | 432 | 1 | 345 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam17189 | Glyco_hydro_30C | 4.18e-08 | 438 | 501 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
| pfam02057 | Glyco_hydro_59 | 0.001 | 168 | 365 | 79 | 236 | Glycosyl hydrolase family 59. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85385.1 | 1.33e-241 | 44 | 503 | 20 | 479 |
| QNT65067.1 | 1.92e-240 | 6 | 504 | 10 | 514 |
| AHW59378.1 | 4.19e-187 | 16 | 503 | 21 | 497 |
| ALJ44501.1 | 3.92e-186 | 39 | 506 | 30 | 492 |
| QUT73412.1 | 3.92e-186 | 39 | 506 | 30 | 492 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5NGK_A | 5.98e-187 | 39 | 506 | 31 | 493 | Theendo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
| 2WNW_A | 9.29e-37 | 52 | 503 | 7 | 442 | Thecrystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2WNW_B The crystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 4FMV_A | 7.48e-07 | 76 | 506 | 6 | 356 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| 4UQA_A | 7.29e-06 | 181 | 473 | 99 | 354 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8J0I9 | 1.16e-37 | 37 | 502 | 23 | 486 | Endo-1,6-beta-D-glucanase BGN16.3 OS=Trichoderma harzianum OX=5544 PE=1 SV=1 |
| O16580 | 2.25e-36 | 51 | 474 | 61 | 483 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
| Q9UB00 | 1.40e-35 | 43 | 472 | 56 | 483 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
| Q4WBR2 | 1.76e-35 | 42 | 502 | 22 | 484 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
| O16581 | 5.00e-33 | 43 | 472 | 54 | 479 | Putative glucosylceramidase 2 OS=Caenorhabditis elegans OX=6239 GN=gba-2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
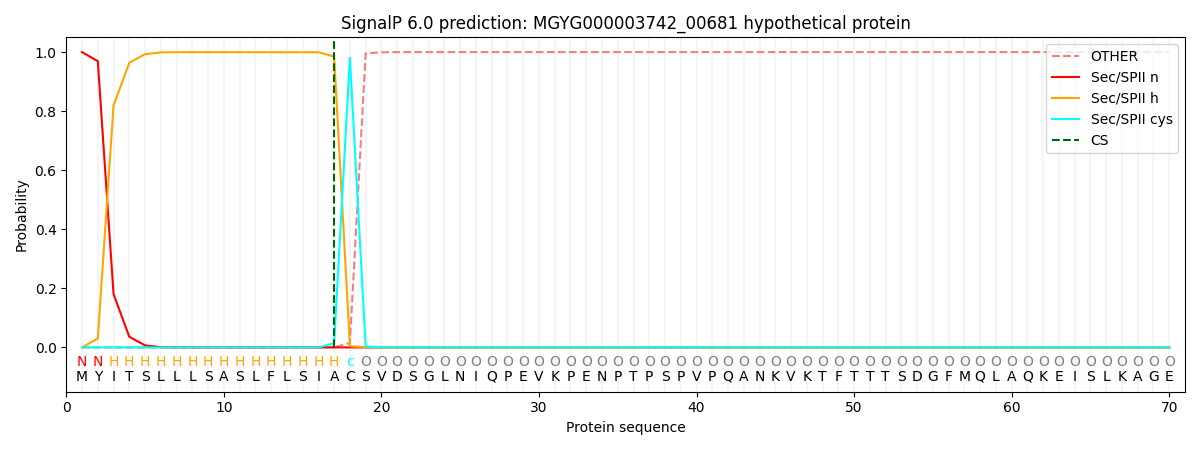
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000139 | 0.999903 | 0.000000 | 0.000000 | 0.000000 |
