You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003744_01203
You are here: Home > Sequence: MGYG000003744_01203
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; | |||||||||||
| CAZyme ID | MGYG000003744_01203 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 679; End: 2265 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 220 | 327 | 9e-17 | 0.7903225806451613 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.26e-18 | 216 | 339 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam16389 | DUF4998 | 9.12e-18 | 26 | 194 | 25 | 199 | Domain of unknown function. This family around 200 residues locates in the N-terminal of some uncharacterized proteins in various Bacteroides and Parabacteroides species. The function of this family remains unknown. |
| pfam16405 | DUF5013 | 2.19e-12 | 360 | 501 | 2 | 145 | Domain of unknown function (DUF5013). This small family of proteins is functionally uncharacterized. This family is found in various Bacteroides and Parabacteroides species. Proteins in this family are around 400 amino acids in length. |
| cd00057 | FA58C | 5.41e-10 | 214 | 326 | 12 | 129 | Substituted updates: Jan 31, 2002 |
| cd08666 | APC10-HECTD3 | 0.002 | 218 | 256 | 11 | 49 | APC10-like DOC1 domain of HECTD3, a HECT E3 ubiquitin ligase protein that mediates substrate ubiquitination. This model represents the APC10/DOC1 domain present in HECTD3, a HECT (Homologous to the E6-AP Carboxyl Terminus) E3 ubiquitin ligase protein. HECT E3 ubiquitin ligases mediate substrate ubiquitination (or ubiquitylation), and are a component of the ubiquitin-26S proteasome pathway for selective proteolytic degradation. They also regulate the trafficking of many receptors, channels, transporters and viral proteins. HECTD3 (HECT domain-containing protein3) contains a C-terminal HECT domain with the active site for ubiquitin transfer onto substrates, and an N-terminal APC10/DOC1 domain, which is responsible for substrate recognition and binding. HECTD3 specifically recognizes the Trio-binding protein, Tara (Trio-associated repeat on actin), implicated in regulating actin cytoskeletal, cell motility and cell growth. Tara also binds to TRF1 and may participate in telomere maintenance and/or mitotic regulation through interacting with TRF1. HECTD3 interacts with and promotes the ubiquitination of Syntaxin 8, an endosomal syntaxin proposed to mediate distinct steps of endosomal protein trafficking. HECTD3-mediated Syntaxin 8 degradation has been suggested to contribute to the pathophysiology of neurodegenerative diseases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH16335.1 | 1.16e-146 | 6 | 528 | 13 | 539 |
| QMW86484.1 | 3.38e-11 | 26 | 339 | 44 | 385 |
| BCA48665.1 | 1.89e-10 | 26 | 339 | 44 | 385 |
| AIE86518.1 | 5.89e-10 | 214 | 340 | 1111 | 1240 |
| QUT41988.1 | 9.96e-10 | 26 | 339 | 33 | 374 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
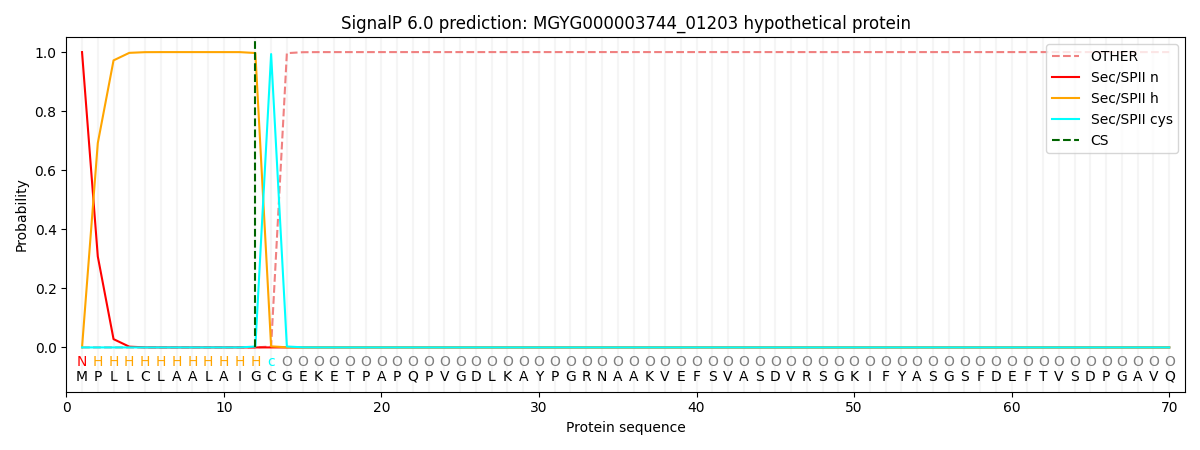
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000039 | 0.000000 | 0.000000 | 0.000000 |
