You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003761_00564
You are here: Home > Sequence: MGYG000003761_00564
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
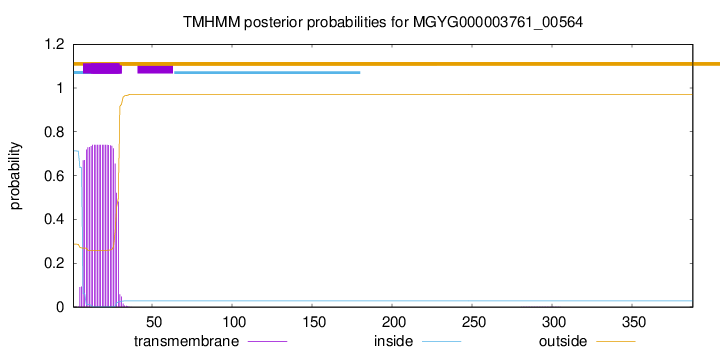
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia_S | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_S | |||||||||||
| CAZyme ID | MGYG000003761_00564 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | Chitin-binding protein CbpD | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6449; End: 7615 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 28 | 207 | 8.1e-52 | 0.9887640449438202 |
| CBM73 | 339 | 385 | 4.1e-17 | 0.8333333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21177 | LPMO_AA10 | 1.33e-77 | 28 | 207 | 1 | 179 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| COG3397 | COG3397 | 5.45e-75 | 21 | 308 | 23 | 305 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
| pfam03067 | LPMO_10 | 8.35e-70 | 28 | 207 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| PRK13211 | PRK13211 | 1.65e-68 | 21 | 386 | 17 | 475 | N-acetylglucosamine-binding protein GbpA. |
| pfam18416 | GbpA_2 | 1.42e-22 | 220 | 316 | 1 | 102 | N-acetylglucosamine binding protein domain 2. This domain can be found in N-acetylglucosamine binding protein (GbpA) from Vibrio cholerae, a bacterial pathogen that colonizes the chitinous exoskeleton of zooplankton as well as the human gastrointestinal tract. GbpA binds to GlcNAc oligosaccharides. Structural comparison show that there are distant structural similarities between domain 2 of GbpA and the beta-domain of the flagellin protein p5. It is suggested that this domain interacts with the bacterial surface, and functions to project an alginate binding domain of the protein from the cell surface. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEF35444.1 | 2.82e-281 | 1 | 388 | 1 | 389 |
| CCH14567.1 | 2.82e-281 | 1 | 388 | 1 | 389 |
| QQQ42318.1 | 5.69e-281 | 1 | 388 | 1 | 389 |
| QGM03071.1 | 5.91e-281 | 1 | 388 | 1 | 390 |
| QNA97174.1 | 8.08e-281 | 1 | 388 | 1 | 389 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6IF7_A | 1.29e-58 | 28 | 209 | 1 | 182 | CrystalStructure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta [Tectaria macrodonta] |
| 5FJQ_A | 5.86e-40 | 28 | 209 | 1 | 179 | Structuraland functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus [Cellvibrio japonicus],5FJQ_B Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus [Cellvibrio japonicus],5FJQ_C Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus [Cellvibrio japonicus] |
| 2XWX_A | 8.89e-32 | 87 | 319 | 56 | 294 | Vibriocholerae colonization factor GbpA crystal structure [Vibrio cholerae],2XWX_B Vibrio cholerae colonization factor GbpA crystal structure [Vibrio cholerae] |
| 5OPF_A | 9.49e-30 | 28 | 209 | 1 | 193 | Structureof LPMO10B from from Micromonospora aurantiaca [Micromonospora aurantiaca ATCC 27029] |
| 4OY7_A | 1.01e-28 | 28 | 212 | 1 | 196 | Structureof cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_B Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_C Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_D Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_E Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_F Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_G Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_H Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02I11 | 9.95e-130 | 19 | 385 | 17 | 386 | Chitin-binding protein CbpD OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=cpbD PE=1 SV=1 |
| Q9I589 | 1.41e-129 | 19 | 385 | 17 | 386 | Chitin-binding protein CbpD OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=cbpD PE=1 SV=1 |
| Q9KLD5 | 6.93e-37 | 87 | 385 | 79 | 482 | GlcNAc-binding protein A OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=gbpA PE=1 SV=1 |
| C3LW75 | 9.57e-37 | 87 | 385 | 79 | 482 | GlcNAc-binding protein A OS=Vibrio cholerae serotype O1 (strain M66-2) OX=579112 GN=gbpA PE=3 SV=1 |
| A5F0H4 | 9.57e-37 | 87 | 385 | 79 | 482 | GlcNAc-binding protein A OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=gbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
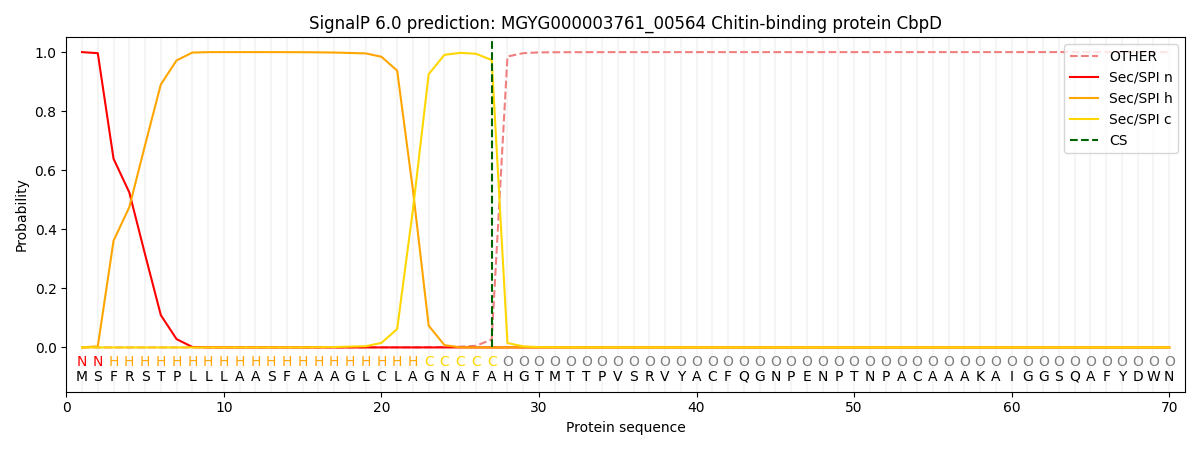
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000268 | 0.999086 | 0.000149 | 0.000179 | 0.000136 | 0.000127 |