You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003769_00399
You are here: Home > Sequence: MGYG000003769_00399
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
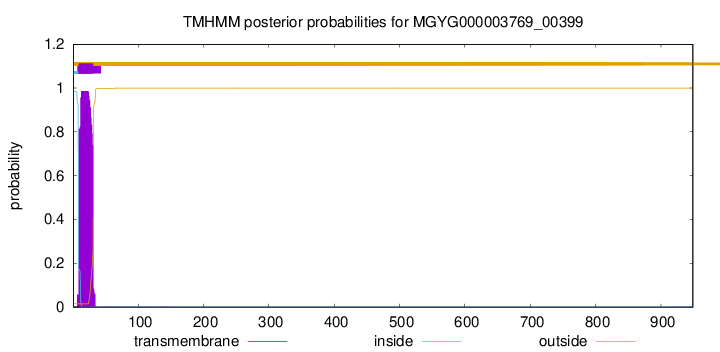
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; | |||||||||||
| CAZyme ID | MGYG000003769_00399 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31585; End: 34434 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 45 | 419 | 2.8e-87 | 0.9493243243243243 |
| GH18 | 656 | 937 | 1.7e-23 | 0.9594594594594594 |
| CBM5 | 478 | 518 | 3.9e-16 | 0.95 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 4.92e-143 | 49 | 416 | 2 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| smart00636 | Glyco_18 | 6.61e-126 | 47 | 416 | 1 | 334 | Glyco_18 domain. |
| COG3325 | ChiA | 9.17e-125 | 45 | 432 | 37 | 439 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 1.61e-102 | 657 | 943 | 2 | 311 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| pfam00704 | Glyco_hydro_18 | 2.53e-98 | 47 | 416 | 1 | 307 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDU53443.1 | 2.02e-193 | 41 | 943 | 52 | 983 |
| QOS39875.1 | 1.15e-189 | 25 | 948 | 32 | 915 |
| QAV17876.1 | 5.59e-167 | 13 | 948 | 24 | 1084 |
| SDT55294.1 | 1.02e-162 | 41 | 943 | 51 | 909 |
| QNU66099.1 | 6.73e-152 | 13 | 440 | 14 | 430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1ITX_A | 1.98e-117 | 42 | 417 | 8 | 407 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6BT9_A | 4.23e-112 | 35 | 416 | 25 | 448 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
| 3EBV_A | 6.80e-111 | 655 | 949 | 4 | 295 | Crystalstructure of putative Chitinase A from Streptomyces coelicolor. [Streptomyces coelicolor] |
| 6KST_A | 4.83e-105 | 46 | 429 | 7 | 370 | Crystalstructure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KST_B Crystal structure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KXL_A Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXL_B Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 6KXM_A | 2.60e-104 | 46 | 429 | 7 | 370 | Crystalstructure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXM_B Crystal structure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 1.61e-113 | 42 | 442 | 40 | 467 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P48827 | 7.20e-71 | 51 | 419 | 43 | 386 | Endochitinase 42 OS=Trichoderma harzianum OX=5544 GN=chit42 PE=1 SV=1 |
| A6N6J0 | 1.21e-70 | 25 | 430 | 18 | 406 | Endochitinase 46 OS=Trichoderma harzianum OX=5544 GN=chit46 PE=1 SV=1 |
| P32470 | 9.55e-70 | 51 | 430 | 43 | 399 | Chitinase 1 OS=Aphanocladium album OX=12942 GN=CHI1 PE=1 SV=2 |
| E9ERT9 | 1.36e-69 | 51 | 430 | 44 | 400 | Endochitinase 1 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chit1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
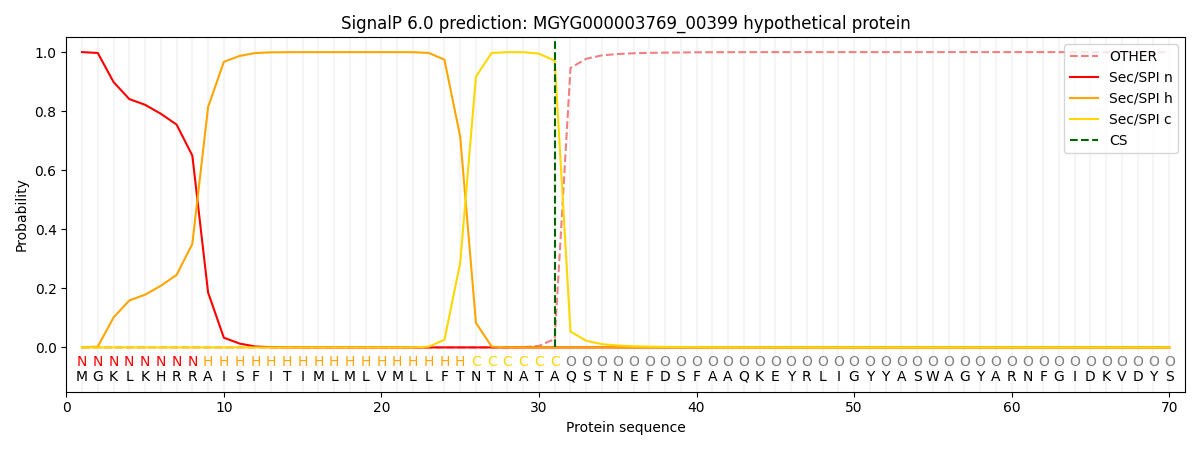
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000911 | 0.997877 | 0.000528 | 0.000274 | 0.000211 | 0.000186 |