You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003769_01484
You are here: Home > Sequence: MGYG000003769_01484
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
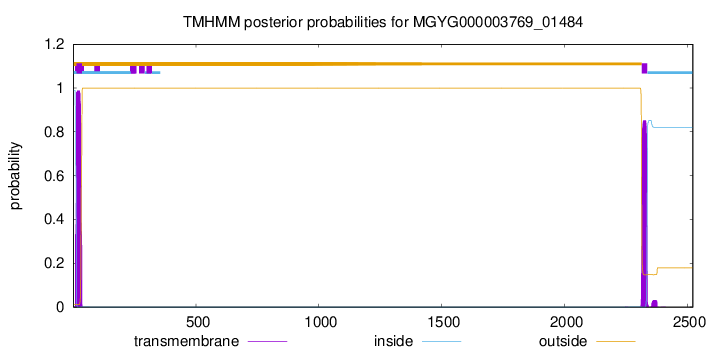
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; | |||||||||||
| CAZyme ID | MGYG000003769_01484 | |||||||||||
| CAZy Family | CBM3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1842; End: 9413 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM3 | 700 | 774 | 2.4e-25 | 0.9090909090909091 |
| CBM3 | 892 | 971 | 1.9e-22 | 0.9659090909090909 |
| CBM3 | 1060 | 1134 | 8.3e-16 | 0.8181818181818182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3209 | RhsA | 7.49e-48 | 1235 | 1919 | 3 | 677 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| COG3209 | RhsA | 7.72e-46 | 1444 | 2138 | 1 | 660 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| COG3209 | RhsA | 5.11e-42 | 1276 | 1976 | 2 | 650 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| COG3209 | RhsA | 1.15e-40 | 1675 | 2354 | 1 | 692 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| COG3209 | RhsA | 2.81e-38 | 1402 | 2020 | 1 | 648 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCN29431.1 | 0.0 | 5 | 1251 | 7 | 1186 |
| AZS13822.1 | 1.44e-87 | 6 | 1251 | 2 | 1043 |
| BCJ98209.1 | 1.88e-65 | 687 | 1036 | 64 | 371 |
| ACM60954.1 | 7.21e-64 | 700 | 1211 | 395 | 935 |
| CDM70444.1 | 3.16e-55 | 886 | 1219 | 1 | 333 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZQW_A | 9.02e-35 | 687 | 822 | 6 | 139 | ChainA, CELLULOSOMAL SCAFFOLDIN [Acetivibrio cellulolyticus],3ZU8_A Chain A, Cellulosomal Scaffoldin [Acetivibrio cellulolyticus],3ZUC_A Chain A, CELLULOSOMAL SCAFFOLDIN [Acetivibrio cellulolyticus] |
| 1NBC_A | 3.02e-31 | 886 | 1032 | 2 | 154 | ChainA, Cellulosomal Scaffolding Protein A [Acetivibrio thermocellus],1NBC_B Chain B, Cellulosomal Scaffolding Protein A [Acetivibrio thermocellus] |
| 4B9F_A | 5.13e-31 | 886 | 1030 | 2 | 152 | ChainA, Cellulosomal-scaffolding Protein A [Acetivibrio thermocellus ATCC 27405],4B9F_B Chain B, Cellulosomal-scaffolding Protein A [Acetivibrio thermocellus ATCC 27405] |
| 1G43_A | 6.53e-31 | 882 | 1030 | 2 | 157 | CrystalStructure Of A Family Iiia Cbd From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum] |
| 4JO5_A | 8.54e-31 | 886 | 1032 | 19 | 171 | ChainA, Cellulosome anchoring protein cohesin region [Acetivibrio thermocellus YS] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22534 | 1.99e-48 | 700 | 1032 | 717 | 1057 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P22533 | 6.81e-48 | 700 | 1032 | 377 | 717 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| Q02934 | 1.28e-38 | 888 | 1032 | 741 | 886 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| P23659 | 1.14e-36 | 883 | 1030 | 837 | 983 | Endoglucanase Z OS=Thermoclostridium stercorarium OX=1510 GN=celZ PE=1 SV=1 |
| P50900 | 3.97e-34 | 686 | 820 | 766 | 896 | Exoglucanase-2 OS=Thermoclostridium stercorarium (strain ATCC 35414 / DSM 8532 / NCIMB 11754) OX=1121335 GN=celY PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
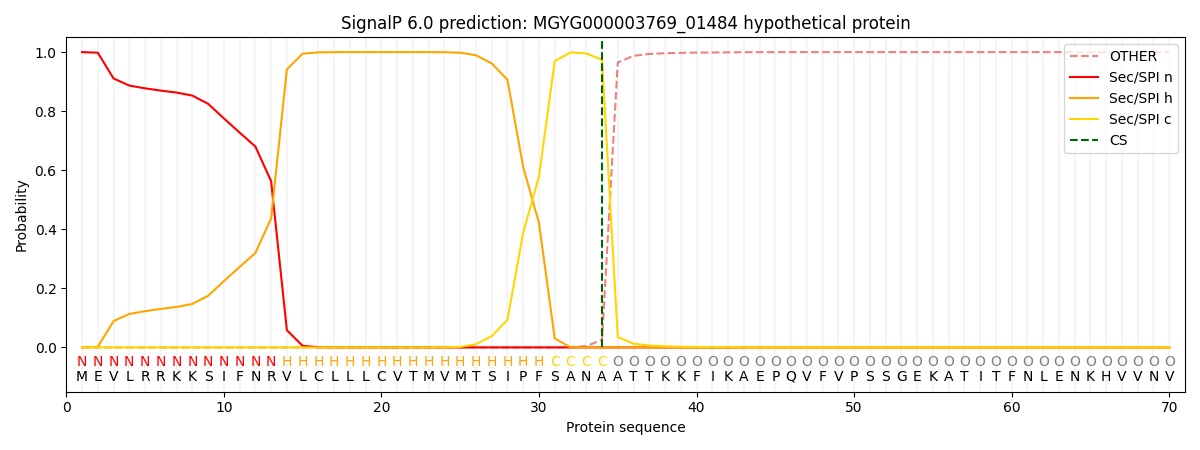
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000308 | 0.998956 | 0.000202 | 0.000198 | 0.000160 | 0.000138 |