You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003769_01513
You are here: Home > Sequence: MGYG000003769_01513
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; | |||||||||||
| CAZyme ID | MGYG000003769_01513 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase E1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42756; End: 44162 Strand: - | |||||||||||
Full Sequence Download help
| MKKIKRMIST LMILIIFFSS GYFKSEQKVI AADTNNDDWL HCVGNKIYDM YGNEVWLTGA | 60 |
| NWFGFNCSEN VFHGAWYDVK ETLTNIANRG IGFLRVPIST ELLYSWMIGK PNKVSSVTAV | 120 |
| NNPPYYVCNP DFYDTVNNKV KNSMEIFDII MGYCKELGIK VMVDVHSPDA NNSGHNYPLW | 180 |
| YGLTTTTAGE ITTEKWIDTL AWLADKYKND DTILAFDIKN EPHGKRGYSP STPSDMAMWD | 240 |
| NTTDENNWKY AAERCARAIL SKNPKVLIMI EGVEQYPKTE LGYSYDTPDI WGASGDASPW | 300 |
| NGAWWGGNLR GVKDYPVDIG TLNSQIVYSP HDYGPSVYSQ SWFDKDFTTQ TLLDDYWYDT | 360 |
| WAYINDQGIA PLFIGEWGGF MDGAKNQKWM TLLRDYMVNN RIHHTFWCIN PNSGDTGGLL | 420 |
| SYDWRTWDEE KYALLKPALW QANGKFIGLD HKIPLGENGI SLGEYYGN | 468 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 49 | 418 | 8.6e-117 | 0.9967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 7.78e-43 | 48 | 413 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.58e-34 | 44 | 446 | 34 | 398 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX41541.1 | 0.0 | 1 | 467 | 1 | 467 |
| BAI52933.1 | 5.26e-306 | 5 | 468 | 3 | 467 |
| BCJ94038.1 | 1.64e-303 | 2 | 468 | 5 | 472 |
| ACL75118.1 | 5.84e-303 | 5 | 468 | 3 | 467 |
| AAG45162.1 | 5.84e-303 | 5 | 468 | 3 | 467 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1ECE_A | 3.23e-69 | 39 | 437 | 5 | 349 | AcidothermusCellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
| 1VRX_A | 2.50e-68 | 39 | 437 | 5 | 349 | ChainA, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
| 3VVG_A | 2.10e-53 | 55 | 436 | 28 | 373 | TheCrystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 3W6M_A | 2.10e-53 | 55 | 436 | 28 | 373 | Contributionof disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 2ZUM_A | 3.59e-53 | 13 | 436 | 10 | 406 | FunctionalAnalysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_A Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_B Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_C Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05332 | 6.68e-297 | 1 | 467 | 1 | 473 | Endoglucanase G OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celG PE=3 SV=1 |
| P10474 | 1.36e-161 | 33 | 468 | 623 | 1029 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P04956 | 1.10e-137 | 36 | 464 | 38 | 469 | Endoglucanase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celB PE=3 SV=1 |
| P50400 | 1.33e-134 | 31 | 463 | 37 | 438 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
| P23548 | 3.76e-72 | 21 | 435 | 19 | 378 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
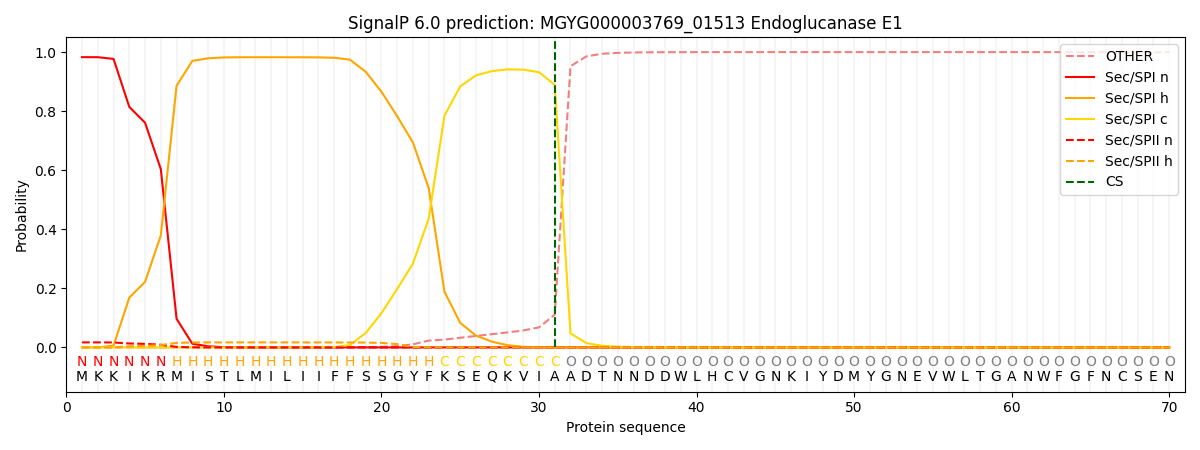
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001032 | 0.979702 | 0.018436 | 0.000335 | 0.000247 | 0.000207 |
