You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003769_01817
You are here: Home > Sequence: MGYG000003769_01817
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
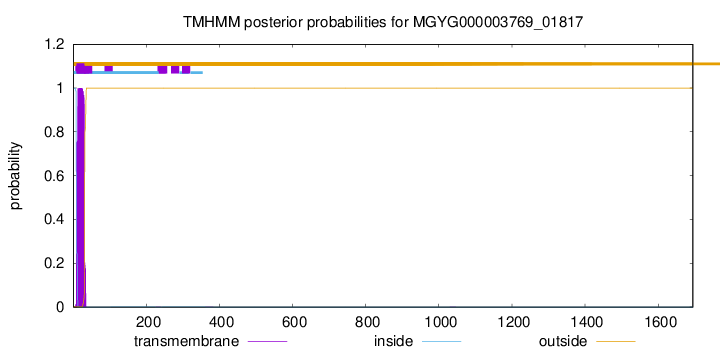
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; | |||||||||||
| CAZyme ID | MGYG000003769_01817 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 40759; End: 45846 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 945 | 1325 | 1.3e-107 | 0.9786666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14592 | Chondroitinas_B | 2.42e-13 | 963 | 1237 | 2 | 328 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
| cd14251 | PL-6 | 4.43e-11 | 963 | 1119 | 2 | 142 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam05887 | Trypan_PARP | 1.52e-07 | 1351 | 1406 | 53 | 108 | Procyclic acidic repetitive protein (PARP). This family consists of several Trypanosoma brucei procyclic acidic repetitive protein (PARP) like sequences. The procyclic acidic repetitive protein (parp) genes of Trypanosoma brucei encode a small family of abundant surface proteins whose expression is restricted to the procyclic form of the parasite. They are found at two unlinked loci, parpA and parpB; transcription of both loci is developmentally regulated. |
| NF033845 | MSCRAMM_ClfB | 1.51e-06 | 1355 | 1397 | 552 | 594 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| PRK05641 | PRK05641 | 4.02e-05 | 1331 | 1420 | 22 | 107 | putative acetyl-CoA carboxylase biotin carboxyl carrier protein subunit; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX44216.1 | 3.59e-129 | 300 | 1348 | 121 | 1206 |
| QEH68113.1 | 4.55e-123 | 464 | 1334 | 277 | 1146 |
| ADZ82419.1 | 7.65e-122 | 464 | 1334 | 277 | 1146 |
| BCJ94249.1 | 1.31e-116 | 396 | 1675 | 217 | 1613 |
| QJS18268.1 | 3.57e-109 | 640 | 1319 | 906 | 1600 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 4.01e-43 | 932 | 1326 | 1 | 373 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 4.13e-58 | 650 | 1226 | 84 | 643 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 3.65e-45 | 930 | 1326 | 24 | 398 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 2.09e-42 | 930 | 1326 | 24 | 398 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
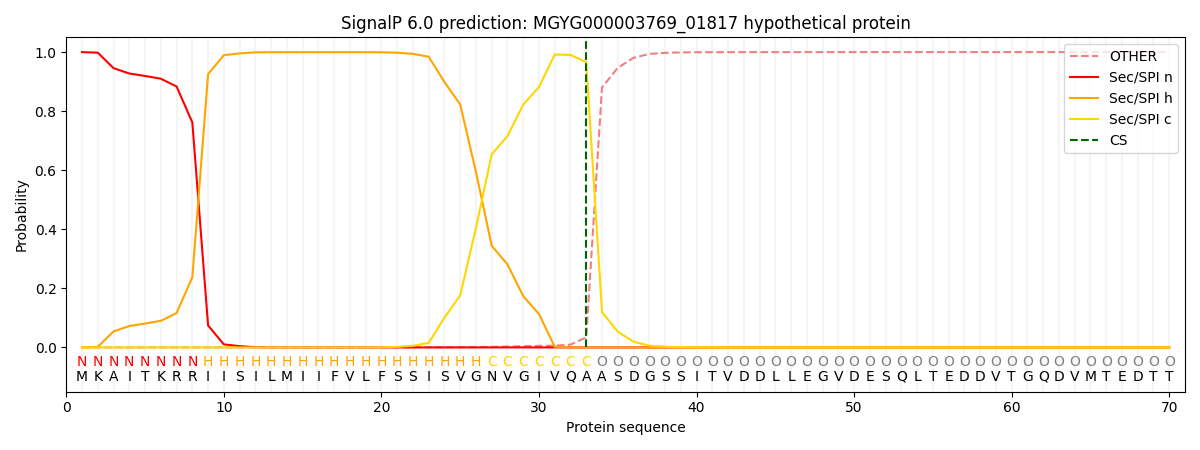
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000442 | 0.998780 | 0.000274 | 0.000179 | 0.000165 | 0.000142 |