You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003769_03127
You are here: Home > Sequence: MGYG000003769_03127
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
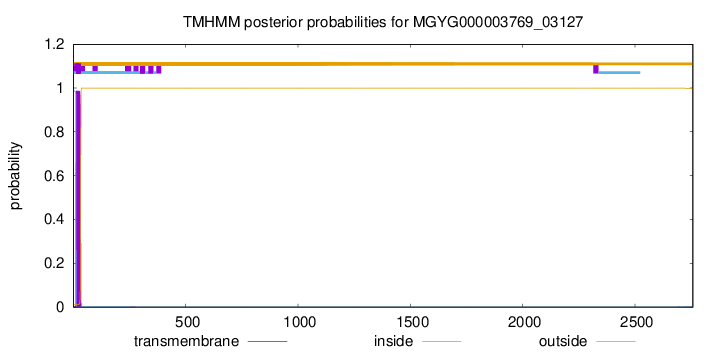
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; | |||||||||||
| CAZyme ID | MGYG000003769_03127 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6697; End: 14973 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 1879 | 2259 | 4.8e-128 | 0.9786666666666667 |
| CE8 | 739 | 1118 | 1.7e-43 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02497 | PLN02497 | 6.74e-18 | 867 | 1124 | 109 | 318 | probable pectinesterase |
| COG4677 | PemB | 1.92e-17 | 682 | 1060 | 33 | 350 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 3.71e-17 | 739 | 1109 | 2 | 275 | Pectinesterase. |
| PLN02745 | PLN02745 | 5.62e-17 | 728 | 1076 | 273 | 527 | Putative pectinesterase/pectinesterase inhibitor |
| PLN02708 | PLN02708 | 1.63e-16 | 725 | 1110 | 229 | 527 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL35206.1 | 0.0 | 144 | 2220 | 113 | 2396 |
| BCJ94249.1 | 0.0 | 1224 | 2757 | 140 | 1641 |
| AFK85374.1 | 0.0 | 167 | 1154 | 749 | 1738 |
| ABX44216.1 | 0.0 | 1296 | 2286 | 205 | 1212 |
| ACR71158.1 | 1.07e-236 | 1312 | 2266 | 612 | 1644 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.30e-42 | 1879 | 2256 | 14 | 369 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 3.90e-17 | 1880 | 2163 | 3 | 325 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 5C1E_A | 8.87e-16 | 739 | 1102 | 11 | 269 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 3.92e-15 | 739 | 1102 | 11 | 269 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 3UW0_A | 2.01e-09 | 734 | 1049 | 30 | 274 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 1.11e-57 | 1495 | 2165 | 23 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A7 | 1.25e-41 | 1879 | 2256 | 39 | 394 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 1.25e-41 | 1879 | 2256 | 39 | 394 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| A2QK82 | 9.72e-15 | 739 | 1102 | 39 | 297 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| A1DBT4 | 7.65e-13 | 929 | 1090 | 143 | 284 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
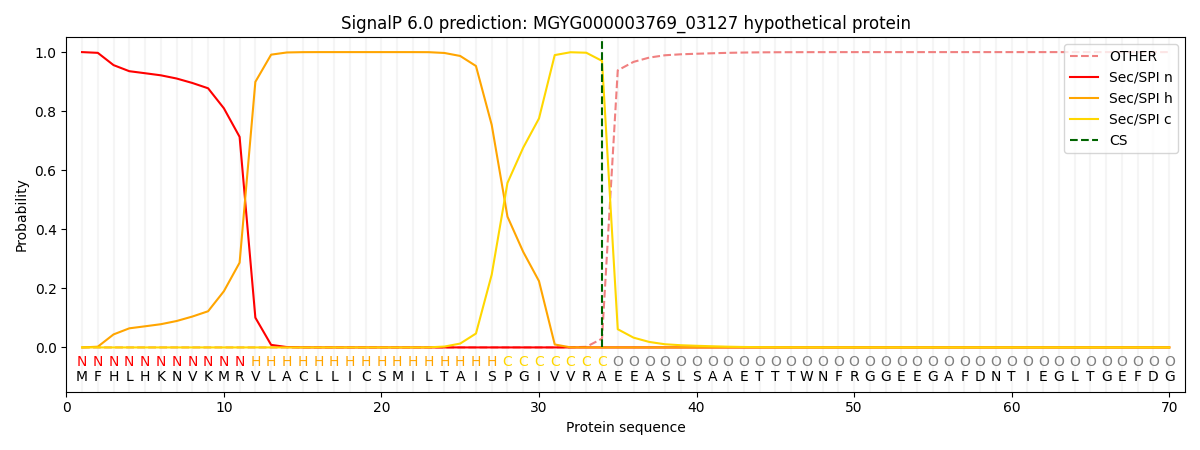
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000255 | 0.999024 | 0.000211 | 0.000177 | 0.000152 | 0.000141 |