You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003786_01181
You are here: Home > Sequence: MGYG000003786_01181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
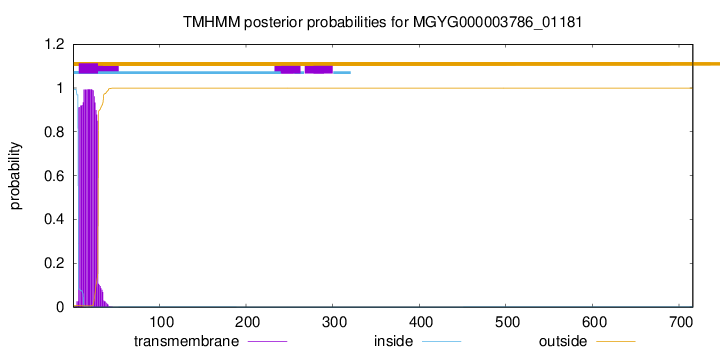
TMHMM annotations
Basic Information help
| Species | Stomatobaculum naviforme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Stomatobaculum; Stomatobaculum naviforme | |||||||||||
| CAZyme ID | MGYG000003786_01181 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21834; End: 23984 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 8.99e-18 | 396 | 553 | 98 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 3.69e-09 | 44 | 99 | 1 | 53 | Bacterial SH3 domain. |
| TIGR04211 | SH3_and_anchor | 2.58e-07 | 47 | 117 | 9 | 77 | SH3 domain protein. Members of this protein family have a signal peptide, a strongly conserved SH3 domain, a variable region, and then a C-terminal hydrophobic transmembrane alpha helix region. |
| COG4991 | YraI | 6.24e-06 | 7 | 101 | 2 | 94 | Uncharacterized conserved protein YraI [Function unknown]. |
| smart00287 | SH3b | 2.15e-05 | 36 | 95 | 1 | 57 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU49989.1 | 1.77e-183 | 2 | 691 | 4 | 705 |
| QQR01104.1 | 1.77e-183 | 2 | 691 | 4 | 705 |
| QIX93811.1 | 8.22e-182 | 2 | 660 | 4 | 683 |
| ASN95383.1 | 5.15e-179 | 2 | 660 | 4 | 689 |
| QRP39925.1 | 5.15e-179 | 2 | 660 | 4 | 689 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 5.46e-11 | 403 | 553 | 97 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 2KQ8_A | 1.30e-07 | 41 | 99 | 7 | 60 | ChainA, Cell wall hydrolase [[Bacillus thuringiensis] serovar konkukian] |
| 4PI7_A | 1.37e-07 | 400 | 539 | 86 | 214 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 8.13e-07 | 400 | 539 | 86 | 214 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q99V41 | 2.43e-09 | 403 | 553 | 1101 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q6GI31 | 2.43e-09 | 403 | 553 | 1110 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
| A7X0T9 | 2.43e-09 | 403 | 553 | 1108 | 1254 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
| Q5HH31 | 2.43e-09 | 403 | 553 | 1109 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain COL) OX=93062 GN=atl PE=3 SV=1 |
| P0C5Z8 | 2.43e-09 | 403 | 553 | 1108 | 1254 | Bifunctional autolysin OS=Staphylococcus aureus OX=1280 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
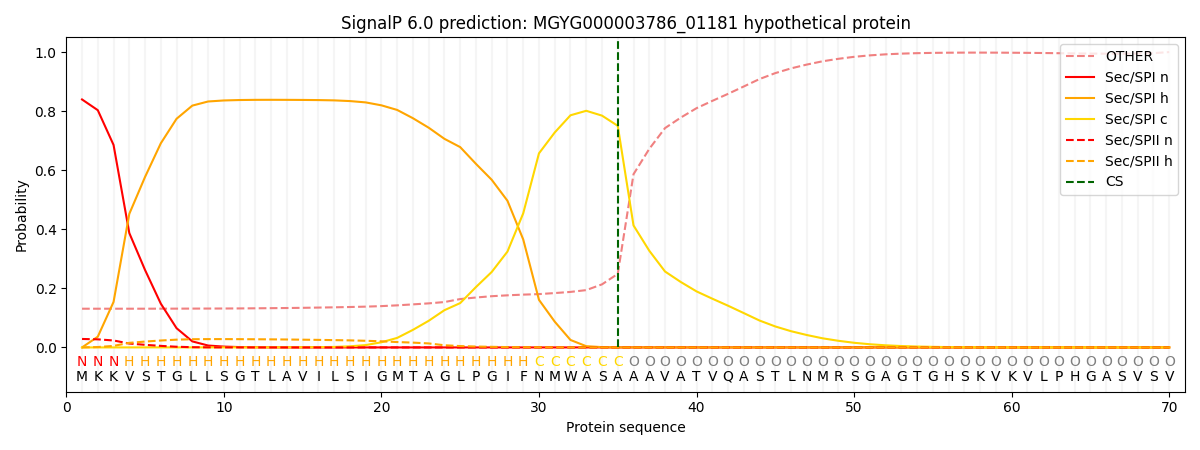
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.141961 | 0.823269 | 0.029850 | 0.003285 | 0.000883 | 0.000732 |