You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003790_00345
You are here: Home > Sequence: MGYG000003790_00345
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
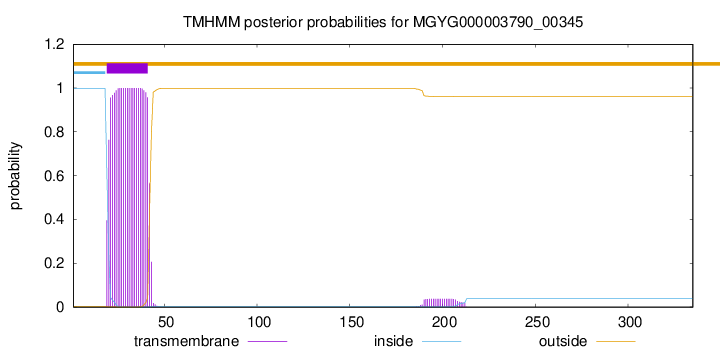
TMHMM annotations
Basic Information help
| Species | Porphyromonas uenonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Porphyromonadaceae; Porphyromonas; Porphyromonas uenonis | |||||||||||
| CAZyme ID | MGYG000003790_00345 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase F | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10637; End: 11644 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 171 | 331 | 5.6e-20 | 0.8592592592592593 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13403 | MLTF-like | 6.81e-67 | 162 | 326 | 1 | 160 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG4623 | MltF | 4.03e-29 | 151 | 318 | 264 | 424 | Membrane-bound lytic murein transglycosylase MltF [Cell wall/membrane/envelope biogenesis, Signal transduction mechanisms]. |
| PRK10859 | PRK10859 | 6.36e-26 | 150 | 315 | 282 | 440 | membrane-bound lytic murein transglycosylase MltF. |
| pfam01464 | SLT | 6.45e-13 | 162 | 269 | 1 | 106 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd16896 | LT_Slt70-like | 9.57e-13 | 163 | 277 | 9 | 121 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE12384.1 | 2.44e-176 | 1 | 334 | 1 | 330 |
| BAK25352.1 | 3.14e-51 | 153 | 328 | 294 | 471 |
| AUR47574.1 | 3.14e-51 | 153 | 328 | 294 | 471 |
| AKV63991.1 | 6.16e-51 | 153 | 328 | 294 | 471 |
| ALO29436.1 | 6.16e-51 | 153 | 328 | 294 | 471 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OZ9_A | 1.30e-14 | 151 | 318 | 252 | 414 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with isoleucine [Pseudomonas aeruginosa PAO1] |
| 4OYV_A | 1.33e-14 | 151 | 318 | 259 | 421 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with leucine [Pseudomonas aeruginosa PAO1] |
| 4OWD_A | 1.33e-14 | 151 | 318 | 259 | 421 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with cysteine [Pseudomonas aeruginosa PAO1],4OXV_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with valine [Pseudomonas aeruginosa PADK2_CF510],4P0G_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with bulgecin and muropeptide [Pseudomonas aeruginosa PAO1],4P11_A Native crystal structure of MltF Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5AA4_B | 1.87e-14 | 151 | 318 | 245 | 407 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA4_D Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013] |
| 5AA4_A | 1.87e-14 | 151 | 318 | 245 | 407 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA4_C Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0AQY9 | 1.68e-18 | 150 | 318 | 281 | 444 | Membrane-bound lytic murein transglycosylase F OS=Maricaulis maris (strain MCS10) OX=394221 GN=mltF PE=3 SV=2 |
| Q1C5E6 | 4.64e-18 | 151 | 318 | 283 | 443 | Membrane-bound lytic murein transglycosylase F OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=mltF PE=3 SV=2 |
| Q74SQ6 | 4.64e-18 | 151 | 318 | 283 | 443 | Membrane-bound lytic murein transglycosylase F OS=Yersinia pestis OX=632 GN=mltF PE=1 SV=1 |
| A7FFU9 | 4.64e-18 | 151 | 318 | 283 | 443 | Membrane-bound lytic murein transglycosylase F OS=Yersinia pseudotuberculosis serotype O:1b (strain IP 31758) OX=349747 GN=mltF PE=3 SV=1 |
| A9R413 | 4.64e-18 | 151 | 318 | 283 | 443 | Membrane-bound lytic murein transglycosylase F OS=Yersinia pestis bv. Antiqua (strain Angola) OX=349746 GN=mltF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
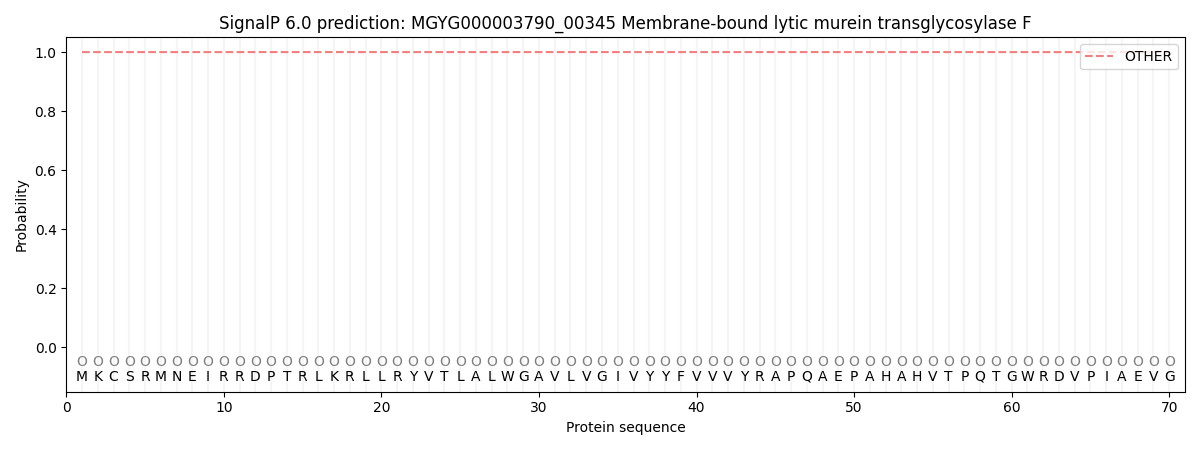
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000045 | 0.000004 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |