You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003792_00289
You are here: Home > Sequence: MGYG000003792_00289
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
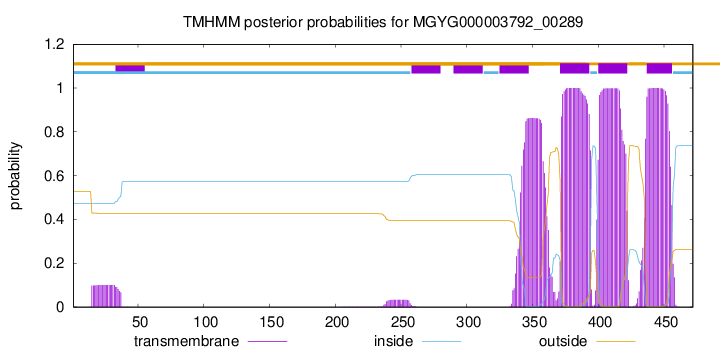
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Brevibacteriaceae; Brevibacterium; | |||||||||||
| CAZyme ID | MGYG000003792_00289 | |||||||||||
| CAZy Family | CE14 | |||||||||||
| CAZyme Description | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10385; End: 11803 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE14 | 5 | 160 | 8.1e-43 | 0.9919354838709677 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03445 | mycothiol_MshB | 1.73e-45 | 5 | 271 | 1 | 262 | N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase. Members of this protein family are N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase, also called 1D-myo-inosityl-2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase, the MshB protein of mycothiol biosynthesis in Mycobacterium tuberculosis and related species. [Cellular processes, Detoxification] |
| COG2120 | LmbE | 6.89e-23 | 3 | 177 | 12 | 143 | N-acetylglucosaminyl deacetylase, LmbE family [Carbohydrate transport and metabolism]. |
| pfam02585 | PIG-L | 3.62e-21 | 6 | 168 | 1 | 124 | GlcNAc-PI de-N-acetylase. Members of this family are related to PIG-L an N-acetylglucosaminylphosphatidylinositol de-N-acetylase (EC:3.5.1.89) that catalyzes the second step in GPI biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIN28729.1 | 1.78e-149 | 4 | 458 | 14 | 492 |
| QCP05414.1 | 2.04e-146 | 4 | 457 | 20 | 505 |
| QQB13329.1 | 3.10e-117 | 4 | 471 | 7 | 447 |
| QQT68206.1 | 3.90e-111 | 4 | 451 | 6 | 434 |
| QPR40189.1 | 3.90e-111 | 4 | 451 | 6 | 434 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EWL_A | 8.86e-28 | 4 | 272 | 7 | 275 | ChainA, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase [Mycobacterium tuberculosis],4EWL_B Chain B, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase [Mycobacterium tuberculosis] |
| 1Q74_A | 9.58e-28 | 4 | 272 | 7 | 275 | ChainA, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) [Mycobacterium tuberculosis],1Q74_B Chain B, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) [Mycobacterium tuberculosis],1Q74_C Chain C, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) [Mycobacterium tuberculosis],1Q74_D Chain D, 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) [Mycobacterium tuberculosis] |
| 1Q7T_A | 1.53e-27 | 4 | 272 | 32 | 300 | ChainA, Rv1170 (MshB) from Mycobacterium tuberculosis [Mycobacterium tuberculosis],1Q7T_B Chain B, Rv1170 (MshB) from Mycobacterium tuberculosis [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D7B6L8 | 4.60e-50 | 5 | 272 | 1 | 261 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase OS=Nocardiopsis dassonvillei (strain ATCC 23218 / DSM 43111 / CIP 107115 / JCM 7437 / KCTC 9190 / NBRC 14626 / NCTC 10488 / NRRL B-5397 / IMRU 509) OX=446468 GN=mshB PE=3 SV=1 |
| D7BQJ3 | 1.19e-47 | 4 | 270 | 10 | 274 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase OS=Streptomyces bingchenggensis (strain BCW-1) OX=749414 GN=mshB PE=3 SV=1 |
| Q82IJ5 | 7.38e-47 | 4 | 270 | 9 | 273 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=mshB PE=3 SV=1 |
| A6W733 | 1.48e-46 | 4 | 270 | 60 | 333 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=mshB PE=3 SV=1 |
| B1W1K9 | 1.50e-46 | 4 | 268 | 9 | 268 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase OS=Streptomyces griseus subsp. griseus (strain JCM 4626 / NBRC 13350) OX=455632 GN=mshB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000042 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |