You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003792_00732
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Brevibacteriaceae; Brevibacterium;
|
| CAZyme ID |
MGYG000003792_00732
|
| CAZy Family |
GT0 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 388 |
|
42940.5 |
8.9181 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003792 |
2144916 |
MAG |
Canada |
North America |
|
| Gene Location |
Start: 4338;
End: 5504
Strand: -
|
No EC number prediction in MGYG000003792_00732.
MGYG000003792_00732 has no CDD domain.
This protein is predicted as OTHER
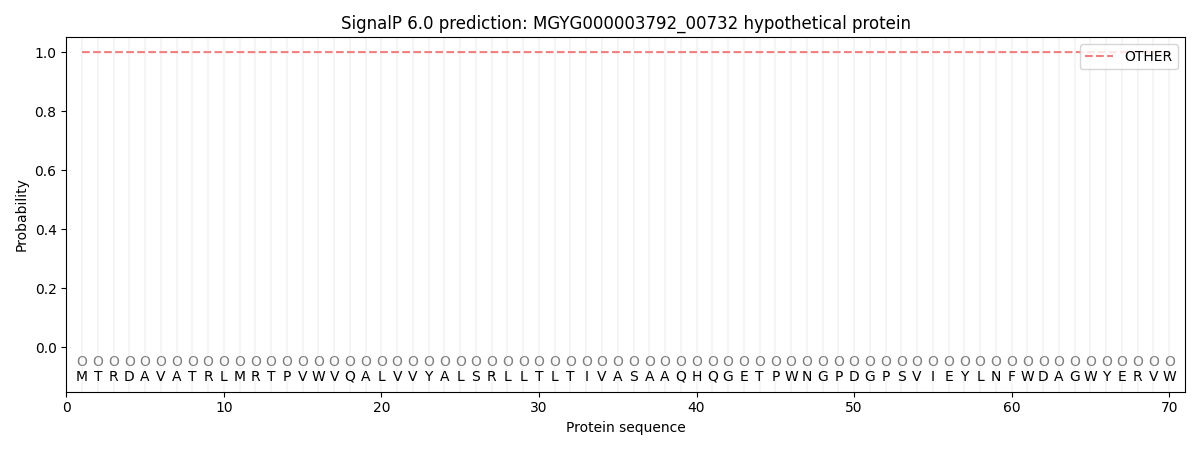
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999998
|
0.000016
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
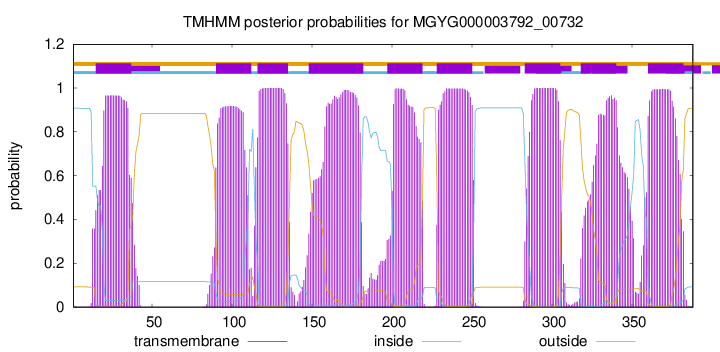
| start |
end |
| 15 |
37 |
| 90 |
112 |
| 116 |
135 |
| 148 |
182 |
| 197 |
219 |
| 228 |
250 |
| 283 |
305 |
| 318 |
340 |
| 360 |
382 |


