You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003803_02558
You are here: Home > Sequence: MGYG000003803_02558
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
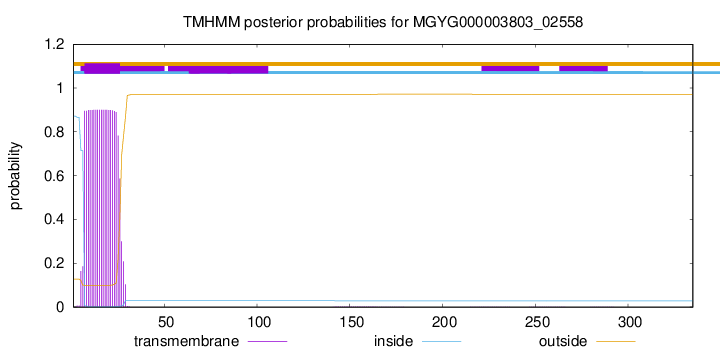
TMHMM annotations
Basic Information help
| Species | Pantoea conspicua | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pantoea; Pantoea conspicua | |||||||||||
| CAZyme ID | MGYG000003803_02558 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Minor endoglucanase Y | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 555; End: 1562 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 1.24e-103 | 21 | 330 | 1 | 318 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 7.38e-56 | 28 | 249 | 29 | 251 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PRK11097 | PRK11097 | 3.41e-40 | 10 | 251 | 7 | 253 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXG54535.1 | 1.54e-226 | 1 | 335 | 1 | 335 |
| QPG27060.1 | 3.11e-226 | 1 | 335 | 1 | 335 |
| QCA06050.1 | 3.11e-226 | 1 | 335 | 1 | 335 |
| ADO11495.1 | 3.11e-226 | 1 | 335 | 1 | 335 |
| AVV36208.1 | 4.42e-226 | 1 | 335 | 1 | 335 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CZL_A | 3.34e-122 | 24 | 335 | 28 | 338 | ChainA, Glucanase [Raoultella ornithinolytica] |
| 5GY3_A | 4.24e-121 | 26 | 335 | 2 | 310 | ChainA, Glucanase [Klebsiella pneumoniae] |
| 1WZZ_A | 5.61e-69 | 22 | 334 | 17 | 331 | Structureof endo-beta-1,4-glucanase CMCax from Acetobacter xylinum [Komagataeibacter xylinus] |
| 6VC5_A | 2.15e-68 | 21 | 330 | 1 | 312 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
| 3QXQ_A | 1.55e-28 | 28 | 245 | 7 | 226 | Structureof the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_B Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_C Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_D Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27032 | 4.64e-147 | 4 | 330 | 3 | 329 | Minor endoglucanase Y OS=Dickeya dadantii (strain 3937) OX=198628 GN=celY PE=1 SV=1 |
| P18336 | 3.00e-115 | 24 | 304 | 23 | 303 | Endoglucanase OS=Cellulomonas uda OX=1714 PE=1 SV=1 |
| P37696 | 3.09e-70 | 22 | 334 | 25 | 339 | Probable endoglucanase OS=Komagataeibacter hansenii OX=436 GN=cmcAX PE=1 SV=1 |
| Q8X5L9 | 1.09e-29 | 1 | 245 | 1 | 247 | Endoglucanase OS=Escherichia coli O157:H7 OX=83334 GN=bcsZ PE=3 SV=1 |
| P37651 | 2.91e-29 | 1 | 245 | 1 | 247 | Endoglucanase OS=Escherichia coli (strain K12) OX=83333 GN=bcsZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
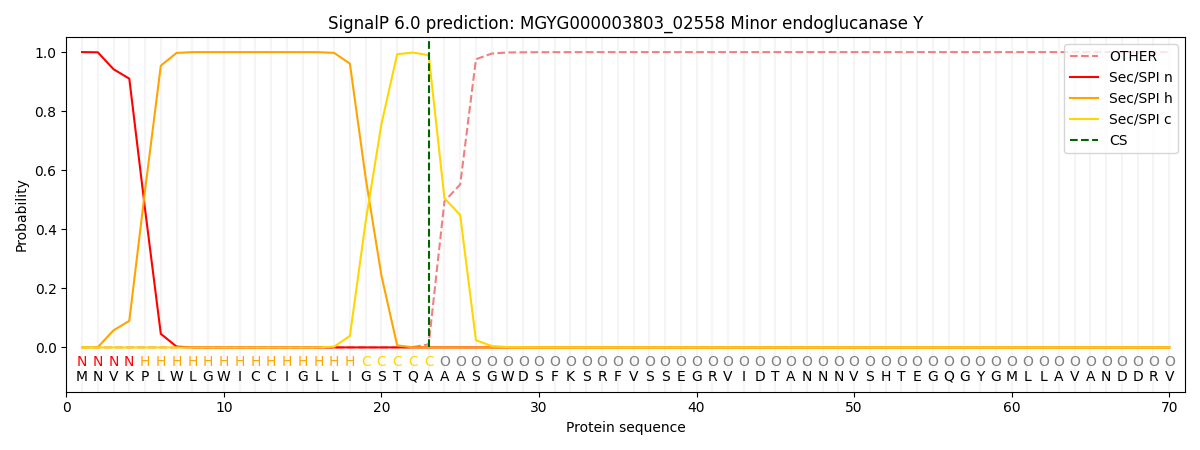
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.999067 | 0.000187 | 0.000145 | 0.000142 | 0.000146 |