You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003831_00512
You are here: Home > Sequence: MGYG000003831_00512
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000003831_00512 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15206; End: 17392 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 438 | 706 | 1.1e-33 | 0.7169230769230769 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00295 | Glyco_hydro_28 | 5.07e-13 | 431 | 674 | 31 | 273 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| COG5434 | Pgu1 | 1.67e-12 | 403 | 727 | 114 | 466 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam17433 | Glyco_hydro_49N | 8.23e-05 | 328 | 411 | 91 | 175 | Glycosyl hydrolase family 49 N-terminal Ig-like domain. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the N-terminal Ig-like fold. |
| PLN03003 | PLN03003 | 8.35e-05 | 434 | 643 | 107 | 291 | Probable polygalacturonase At3g15720 |
| PLN03010 | PLN03010 | 1.25e-04 | 484 | 549 | 167 | 234 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL02017.1 | 1.70e-147 | 16 | 708 | 19 | 721 |
| QRP58902.1 | 1.46e-138 | 19 | 679 | 44 | 725 |
| QQA09745.1 | 1.46e-138 | 19 | 679 | 44 | 725 |
| QQT79214.1 | 1.46e-138 | 19 | 679 | 44 | 725 |
| ASW16403.1 | 1.46e-138 | 19 | 679 | 44 | 725 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1BHE_A | 7.36e-09 | 470 | 697 | 151 | 352 | ChainA, POLYGALACTURONASE [Pectobacterium carotovorum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P18192 | 8.20e-09 | 470 | 697 | 177 | 378 | Endo-polygalacturonase OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=peh PE=3 SV=1 |
| A7PZL3 | 1.83e-08 | 437 | 550 | 167 | 277 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| P26509 | 4.44e-08 | 470 | 697 | 177 | 378 | Endo-polygalacturonase OS=Pectobacterium parmentieri OX=1905730 GN=pehA PE=1 SV=1 |
| P24548 | 1.18e-07 | 444 | 638 | 73 | 261 | Exopolygalacturonase (Fragment) OS=Oenothera organensis OX=3945 PE=2 SV=1 |
| B0YDE8 | 7.96e-07 | 437 | 683 | 132 | 379 | Probable exopolygalacturonase X OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pgaX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
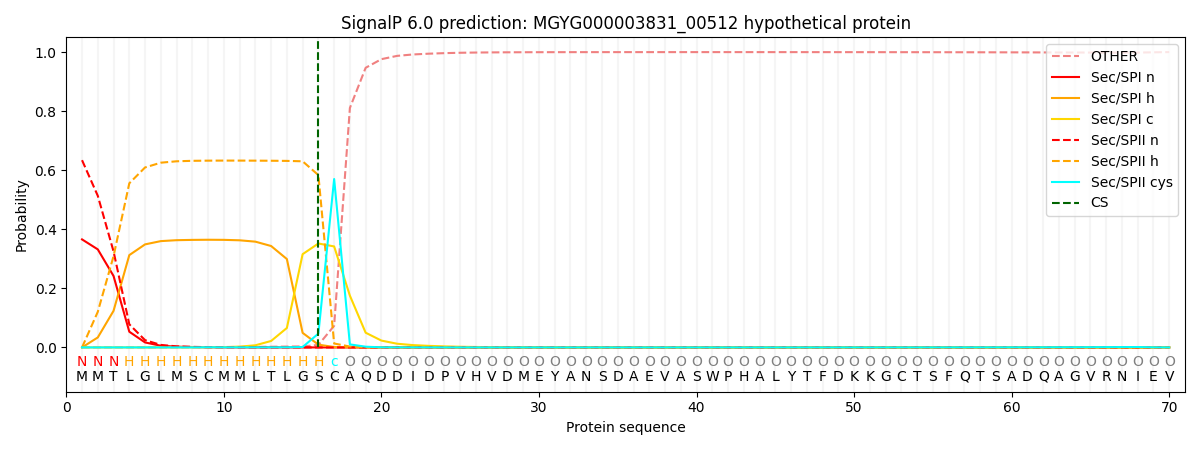
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000683 | 0.358494 | 0.640314 | 0.000230 | 0.000147 | 0.000131 |
