You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003831_01339
You are here: Home > Sequence: MGYG000003831_01339
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000003831_01339 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22688; End: 25861 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 237 | 467 | 1.7e-22 | 0.9419642857142857 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 8.40e-28 | 828 | 978 | 2 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam11790 | Glyco_hydro_cc | 7.77e-15 | 238 | 469 | 22 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| cd00110 | LamG | 2.78e-05 | 828 | 948 | 5 | 124 | Laminin G domain; Laminin G-like domains are usually Ca++ mediated receptors that can have binding sites for steroids, beta1 integrins, heparin, sulfatides, fibulin-1, and alpha-dystroglycans. Proteins that contain LamG domains serve a variety of purposes including signal transduction via cell-surface steroid receptors, adhesion, migration and differentiation through mediation of cell adhesion molecules. |
| cd00063 | FN3 | 0.001 | 596 | 684 | 7 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00560 | LamGL | 0.004 | 845 | 938 | 2 | 97 | LamG-like jellyroll fold domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUC19783.1 | 5.65e-217 | 26 | 1002 | 363 | 1338 |
| APZ45921.1 | 5.65e-217 | 26 | 1002 | 363 | 1338 |
| AXY72824.1 | 1.42e-182 | 32 | 1019 | 391 | 1340 |
| AEV99551.1 | 1.17e-171 | 29 | 980 | 412 | 1336 |
| QGK74225.1 | 1.05e-105 | 16 | 779 | 538 | 1285 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QVS_A | 3.02e-20 | 788 | 980 | 16 | 219 | ChainA, S-layer domain-containing protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
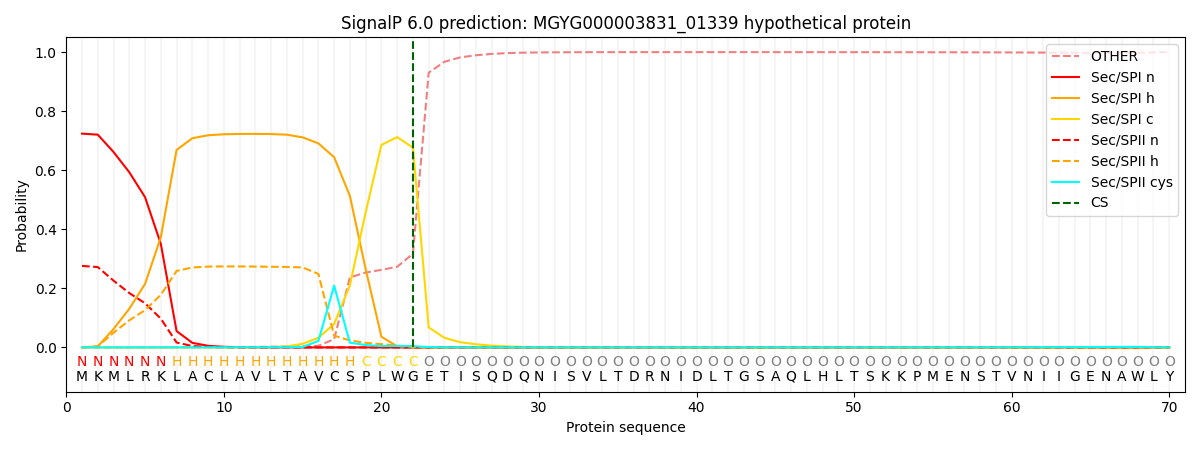
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000801 | 0.712495 | 0.285819 | 0.000324 | 0.000286 | 0.000262 |
