You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003831_01426
You are here: Home > Sequence: MGYG000003831_01426
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
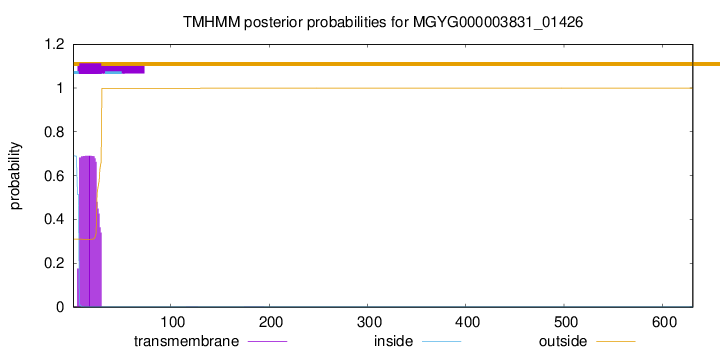
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000003831_01426 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6548; End: 8443 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 49 | 553 | 1.1e-191 | 0.9980119284294234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 0.0 | 49 | 553 | 1 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 6.60e-115 | 34 | 557 | 1 | 505 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| pfam16375 | DUF4986 | 1.50e-06 | 582 | 628 | 38 | 84 | Domain of unknown function. This family around 150 residues locates in the C-terminal of some uncharacterized proteins in various Bacteroides and Bacillus species. The function of this family remains unknown. |
| cd04792 | LanM-like | 1.29e-04 | 184 | 265 | 597 | 689 | Cyclases involved in the biosynthesis of class II lantibiotics, and similar proteins. LanM-like proteins. LanM is a bifunctional enzyme, involved in the synthesis of class II lantibiotics. It is responsible for both the dehydration and the cyclization of the precursor-peptide during lantibiotic synthesis. The C-terminal domain shows similarity to LanC, the cyclase component of the lan operon, but the N terminus seems to be unrelated to the dehydratase, LanB. |
| cd20171 | M34_peptidase | 0.006 | 98 | 163 | 65 | 124 | Peptidase family M34 includes the C-terminal catalytic domain of anthrax lethal factor (ATLF), the protective antigen-binding domains of ATLF and edema factor, and Pro-Pro endopeptidase. Peptidase family M34 (also known as the anthrax lethal factor family) includes the C-terminal catalytic domain of anthrax lethal factor (ATLF, EC 3.4.24.83), and the N-terminal protective antigen-binding domains (PABDs) of ATLF and edema factor (EF). ATLF and EF are enzyme components of anthrax toxin and are carried into the cell by a third component, the protective antigen (PA). ATLF is a highly selective protease whose major substrates are mitogen-activated protein kinase kinases (MKKs). At its N-terminus, ATLF has a PABD domain which lacks the hallmark metalloprotease motif HEXXH, and, at its C-terminus, the related catalytic domain has the HEXXH motif where the two His residues bind a single zinc atom, and the Glu has a catalytic role. EF acts as a Ca2+- and calmodulin-dependent adenylyl cyclase that can cause edema when associated with PA. EF is comprised of the PABD and an adenylyl cyclase domain. This family also includes Pro-Pro endopeptidase (PPEP-1; EC 3.4.24.89, also known as Zmp1) which is an extracellular metalloprotease that shows a unique specificity for hydrolyzing a Pro-Pro bond and is involved in bacterial adhesion. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA49451.1 | 5.38e-295 | 2 | 631 | 5 | 640 |
| ALJ42763.1 | 1.54e-294 | 2 | 631 | 5 | 640 |
| ALJ62034.1 | 2.18e-294 | 3 | 630 | 4 | 638 |
| QUT42773.1 | 3.09e-294 | 2 | 631 | 5 | 640 |
| QQA07126.1 | 4.08e-294 | 2 | 631 | 3 | 638 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YQH_AAA | 1.55e-111 | 35 | 629 | 18 | 649 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
| 5OPJ_A | 7.67e-107 | 35 | 629 | 18 | 649 | Beta-L-arabinofuranosidase[Bacteroides thetaiotaomicron] |
| 6EX6_A | 1.26e-24 | 131 | 551 | 88 | 543 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4QJY_A | 2.35e-19 | 119 | 527 | 83 | 530 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 4QK0_A | 6.78e-18 | 119 | 527 | 83 | 530 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
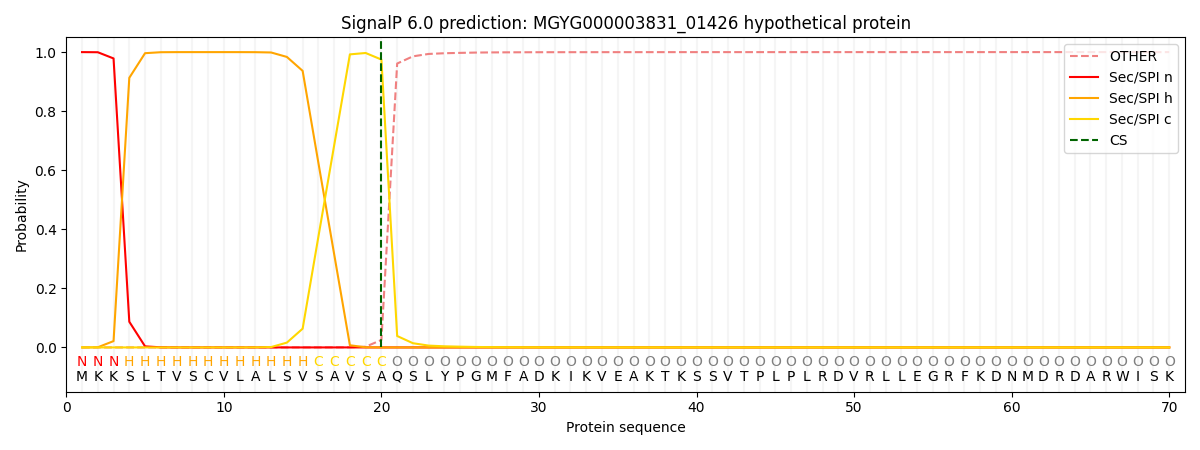
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000342 | 0.998938 | 0.000191 | 0.000178 | 0.000166 | 0.000151 |