You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003846_01667
You are here: Home > Sequence: MGYG000003846_01667
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
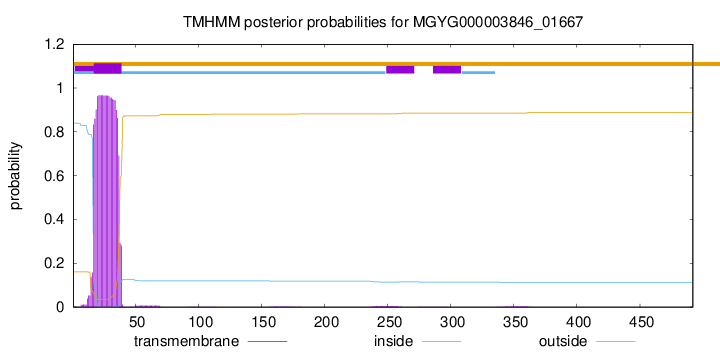
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; | |||||||||||
| CAZyme ID | MGYG000003846_01667 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11857; End: 13335 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 57 | 337 | 1.9e-22 | 0.6981818181818182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.91e-06 | 80 | 327 | 25 | 244 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL08912.1 | 1.96e-114 | 36 | 490 | 32 | 469 |
| BBL11704.1 | 1.96e-114 | 36 | 490 | 32 | 469 |
| AXE20299.1 | 3.34e-114 | 36 | 489 | 29 | 474 |
| QYY35449.1 | 3.47e-114 | 45 | 489 | 15 | 454 |
| BBL00987.1 | 1.56e-113 | 36 | 490 | 32 | 469 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6Z310 | 2.02e-12 | 29 | 359 | 29 | 366 | Putative mannan endo-1,4-beta-mannosidase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN9 PE=2 SV=2 |
| Q9FJZ3 | 2.91e-11 | 30 | 359 | 25 | 362 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
| Q9SG94 | 4.84e-11 | 47 | 360 | 39 | 366 | Mannan endo-1,4-beta-mannosidase 3 OS=Arabidopsis thaliana OX=3702 GN=MAN3 PE=2 SV=1 |
| B8NIV9 | 2.23e-09 | 45 | 245 | 134 | 317 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
| Q2U2I3 | 2.96e-09 | 45 | 245 | 134 | 317 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
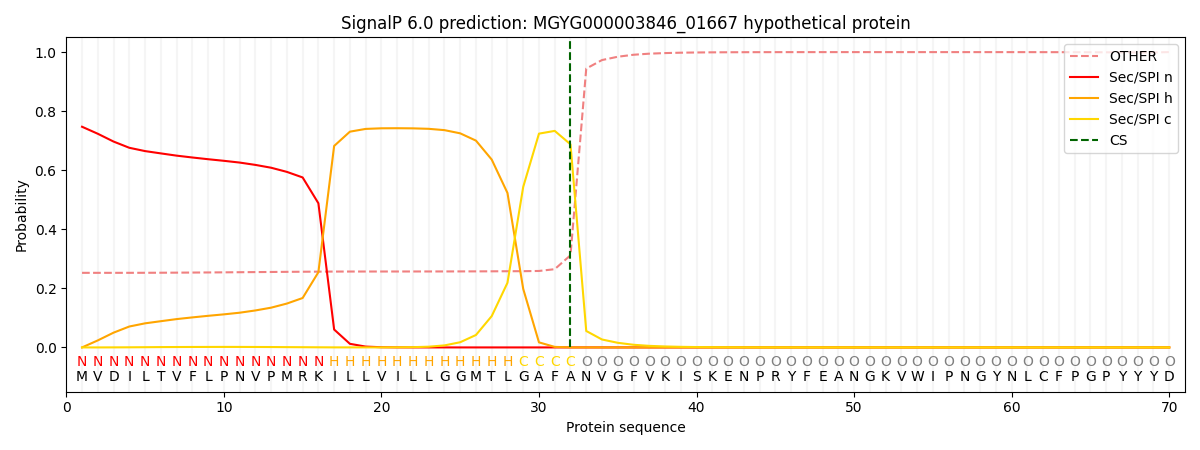
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.262100 | 0.736261 | 0.000644 | 0.000437 | 0.000245 | 0.000282 |