You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003856_01434
You are here: Home > Sequence: MGYG000003856_01434
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paramuribaculum sp900551515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp900551515 | |||||||||||
| CAZyme ID | MGYG000003856_01434 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10749; End: 13283 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 171 | 465 | 4.9e-105 | 0.9932203389830508 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 4.21e-155 | 171 | 450 | 1 | 278 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 3.20e-11 | 51 | 164 | 6 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 9.85e-11 | 625 | 733 | 14 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG3525 | Chb | 9.44e-06 | 49 | 223 | 136 | 311 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
| cd00057 | FA58C | 6.83e-05 | 635 | 733 | 22 | 138 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI62100.1 | 0.0 | 14 | 844 | 22 | 854 |
| BBL05673.1 | 0.0 | 18 | 844 | 22 | 846 |
| AHF11939.1 | 0.0 | 17 | 842 | 23 | 852 |
| ADV43394.1 | 0.0 | 16 | 842 | 21 | 847 |
| ALB75783.1 | 0.0 | 20 | 842 | 27 | 850 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2J4G_A | 5.65e-150 | 89 | 691 | 50 | 667 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482],2J4G_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482] |
| 2JIW_A | 5.82e-150 | 89 | 691 | 51 | 668 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine [Bacteroides thetaiotaomicron VPI-5482],2JIW_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine [Bacteroides thetaiotaomicron VPI-5482] |
| 2J47_A | 5.82e-150 | 89 | 691 | 51 | 668 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with a imidazole-pugnac hybrid inhibitor [Bacteroides thetaiotaomicron VPI-5482],2W4X_A BtGH84 in complex with STZ [Bacteroides thetaiotaomicron VPI-5482],2W66_A BtGH84 in complex with HQ602 [Bacteroides thetaiotaomicron VPI-5482],2W66_B BtGH84 in complex with HQ602 [Bacteroides thetaiotaomicron VPI-5482],2W67_A BtGH84 in complex with FMA34 [Bacteroides thetaiotaomicron VPI-5482],2W67_B BtGH84 in complex with FMA34 [Bacteroides thetaiotaomicron VPI-5482],2WCA_A BtGH84 in complex with n-butyl pugnac [Bacteroides thetaiotaomicron VPI-5482],2XJ7_A BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine [Bacteroides thetaiotaomicron VPI-5482],2XJ7_B BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine [Bacteroides thetaiotaomicron VPI-5482],2XM1_A BtGH84 in complex with N-acetyl gluconolactam [Bacteroides thetaiotaomicron VPI-5482],2XM1_B BtGH84 in complex with N-acetyl gluconolactam [Bacteroides thetaiotaomicron VPI-5482],2XM2_A BtGH84 in complex with LOGNAc [Bacteroides thetaiotaomicron VPI-5482],2XM2_B BtGH84 in complex with LOGNAc [Bacteroides thetaiotaomicron VPI-5482],4UR9_A Structure of ligand bound glycosylhydrolase [Bacteroides thetaiotaomicron],4UR9_B Structure of ligand bound glycosylhydrolase [Bacteroides thetaiotaomicron],5FKY_A Structure of a hydrolase bound with an inhibitor [Bacteroides thetaiotaomicron],5FKY_B Structure of a hydrolase bound with an inhibitor [Bacteroides thetaiotaomicron],5FL0_A Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL0_B Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL1_A Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL1_B Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron] |
| 7K41_A | 6.36e-150 | 89 | 691 | 54 | 671 | ChainA, O-GlcNAcase BT_4395 [Escherichia coli K-12] |
| 5ABE_A | 7.82e-150 | 89 | 691 | 61 | 678 | Structureof GH84 with ligand [Bacteroides thetaiotaomicron],5ABE_B Structure of GH84 with ligand [Bacteroides thetaiotaomicron],5ABF_A Structure of GH84 with ligand [Bacteroides thetaiotaomicron],5ABG_A Structure of GH84 with ligand [Bacteroides thetaiotaomicron],5ABG_B Structure of GH84 with ligand [Bacteroides thetaiotaomicron],5ABH_A Structure of GH84 with ligand [Bacteroides thetaiotaomicron],5ABH_B Structure of GH84 with ligand [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q89ZI2 | 5.91e-149 | 89 | 691 | 72 | 689 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q8XL08 | 6.00e-90 | 116 | 724 | 133 | 746 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 8.29e-90 | 116 | 724 | 133 | 746 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| P26831 | 1.77e-51 | 115 | 747 | 133 | 804 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| O60502 | 1.63e-36 | 172 | 433 | 63 | 331 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
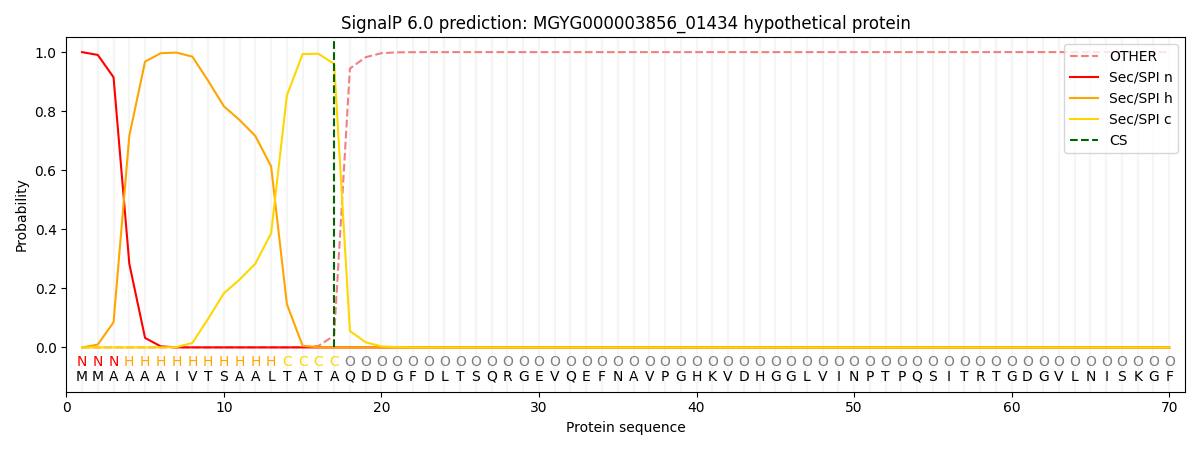
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000666 | 0.998332 | 0.000220 | 0.000291 | 0.000227 | 0.000214 |
