You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003858_01527
You are here: Home > Sequence: MGYG000003858_01527
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
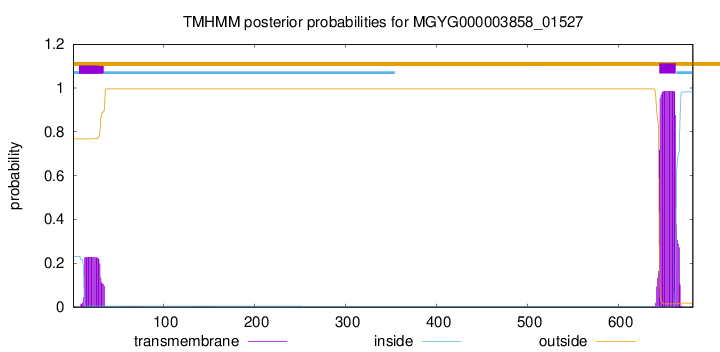
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS363; | |||||||||||
| CAZyme ID | MGYG000003858_01527 | |||||||||||
| CAZy Family | GH44 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7051; End: 9099 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH44 | 41 | 540 | 3.1e-125 | 0.9844357976653697 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12891 | Glyco_hydro_44 | 8.91e-44 | 139 | 317 | 36 | 234 | Glycoside hydrolase family 44. This is a family of bacterial glycoside hydrolases formerly known as cellulase family J, and now known as Cel44A. It is one of the major enzymatic components of the cellulosome of Clostridium thermocellum strain F1 and of many other Firmicutes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMO13192.1 | 4.85e-112 | 43 | 538 | 11 | 514 |
| ADZ19966.1 | 1.14e-111 | 10 | 535 | 1 | 535 |
| AEI34285.1 | 1.14e-111 | 10 | 535 | 1 | 535 |
| AAK78891.1 | 1.14e-111 | 10 | 535 | 1 | 535 |
| AWV80610.1 | 1.14e-111 | 10 | 535 | 1 | 535 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2E0P_A | 4.65e-114 | 43 | 545 | 12 | 518 | ChainA, Endoglucanase [Acetivibrio thermocellus],2E4T_A Chain A, Endoglucanase [Acetivibrio thermocellus],2EO7_A Chain A, Endoglucanase [Acetivibrio thermocellus] |
| 2EEX_A | 1.30e-113 | 43 | 545 | 12 | 518 | ChainA, Endoglucanase [Acetivibrio thermocellus],2EJ1_A Chain A, Endoglucanase [Acetivibrio thermocellus],2EQD_A Chain A, Endoglucanase [Acetivibrio thermocellus] |
| 3IK2_A | 3.41e-113 | 42 | 535 | 4 | 503 | CrystalStructure of a Glycoside Hydrolase Family 44 Endoglucanase produced by Clostridium acetobutylium ATCC 824 [Clostridium acetobutylicum ATCC 824] |
| 2YKK_A | 1.38e-98 | 42 | 535 | 11 | 509 | Structureof a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa],3ZQ9_A Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa] |
| 2YIH_A | 2.09e-97 | 42 | 535 | 11 | 509 | Structureof a Paenibacillus polymyxa Xyloglucanase from GH family 44 with Xyloglucan [Paenibacillus polymyxa],2YJQ_A Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa],2YJQ_B Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 [Paenibacillus polymyxa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29719 | 1.31e-97 | 6 | 554 | 1 | 552 | Endoglucanase A OS=Paenibacillus lautus OX=1401 GN=celA PE=3 SV=1 |
| P22533 | 1.36e-92 | 43 | 512 | 790 | 1273 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
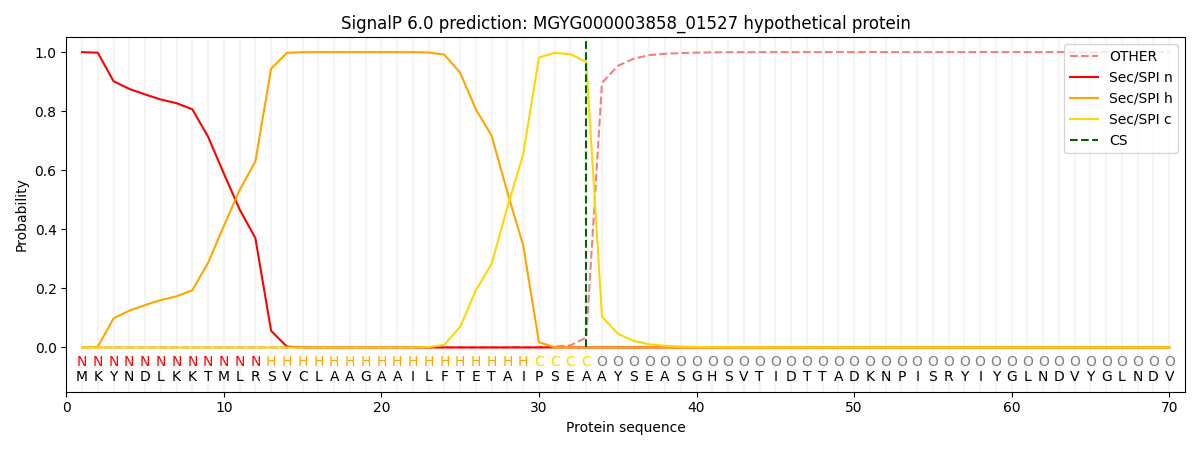
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000293 | 0.998993 | 0.000190 | 0.000188 | 0.000167 | 0.000143 |