You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003878_00208
You are here: Home > Sequence: MGYG000003878_00208
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella oris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella oris | |||||||||||
| CAZyme ID | MGYG000003878_00208 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | Isomalto-dextranase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35586; End: 37037 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 191 | 458 | 3.1e-28 | 0.9388646288209607 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17801 | Melibiase_C | 1.56e-15 | 409 | 479 | 5 | 74 | Alpha galactosidase C-terminal beta sandwich domain. This domain is found at the C-terminus of alpha galactosidase enzymes. |
| PLN02808 | PLN02808 | 1.69e-09 | 360 | 480 | 266 | 384 | alpha-galactosidase |
| PLN02229 | PLN02229 | 4.66e-08 | 338 | 482 | 267 | 420 | alpha-galactosidase |
| COG3669 | AfuC | 0.004 | 38 | 216 | 2 | 191 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH14481.1 | 0.0 | 1 | 483 | 1 | 483 |
| EFV04299.1 | 0.0 | 1 | 483 | 1 | 483 |
| BBA29349.1 | 5.28e-304 | 1 | 481 | 1 | 482 |
| QUB45412.1 | 1.76e-302 | 1 | 481 | 1 | 482 |
| ANR72383.1 | 1.76e-302 | 1 | 481 | 1 | 482 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AWO_A | 7.07e-77 | 53 | 480 | 38 | 466 | Arthrobacterglobiformis T6 isomalto-dextranse [Arthrobacter globiformis],5AWP_A Arthrobacter globiformis T6 isomalto-dextranase complexed with isomaltose [Arthrobacter globiformis],5AWQ_A Arthrobacter globiformis T6 isomalto-dextranse complexed with panose [Arthrobacter globiformis] |
| 4NX0_A | 5.56e-13 | 27 | 482 | 5 | 443 | Crystalstructure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_B Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_C Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_D Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_E Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_F Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_G Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4NX0_H Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44052 | 1.01e-76 | 53 | 480 | 64 | 492 | Isomalto-dextranase OS=Arthrobacter globiformis OX=1665 GN=imd PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
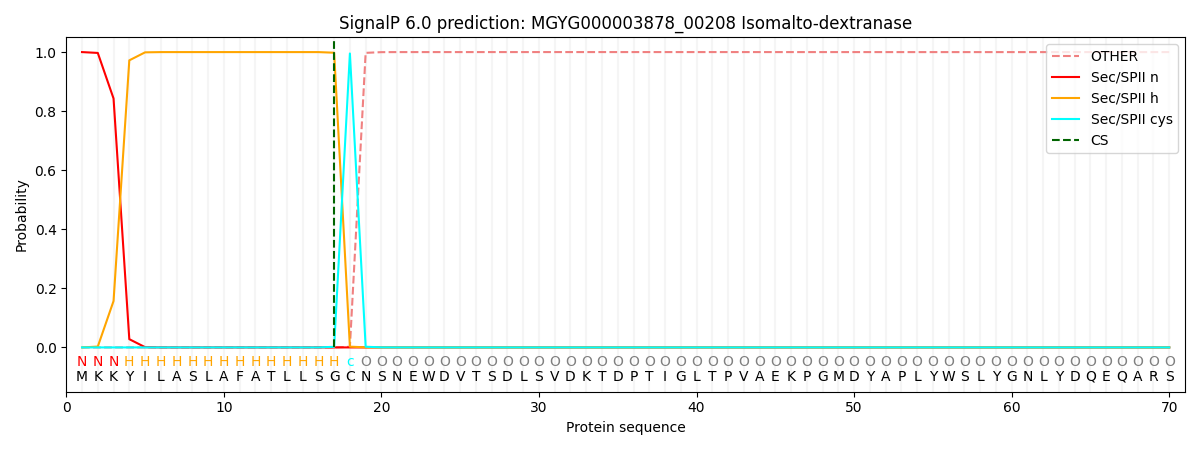
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000047 | 0.000000 | 0.000000 | 0.000000 |
