You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003880_00169
You are here: Home > Sequence: MGYG000003880_00169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900543205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900543205 | |||||||||||
| CAZyme ID | MGYG000003880_00169 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 205891; End: 208128 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 63 | 276 | 9.3e-63 | 0.9490740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 2.35e-104 | 13 | 738 | 18 | 758 | beta-glucosidase BglX. |
| COG1472 | BglX | 3.23e-67 | 100 | 377 | 80 | 365 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 8.82e-59 | 351 | 562 | 1 | 198 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PLN03080 | PLN03080 | 1.81e-54 | 40 | 731 | 56 | 769 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 6.93e-43 | 97 | 317 | 82 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SJX74207.1 | 0.0 | 7 | 745 | 1 | 745 |
| ADB80109.1 | 0.0 | 7 | 745 | 14 | 761 |
| BBE20347.1 | 5.77e-268 | 30 | 745 | 29 | 759 |
| QIA09294.1 | 2.16e-265 | 8 | 745 | 7 | 759 |
| AHW58987.1 | 1.36e-261 | 8 | 745 | 8 | 760 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MS2_A | 5.54e-139 | 36 | 745 | 4 | 668 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 5WAB_A | 2.29e-114 | 44 | 745 | 12 | 663 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 2X40_A | 7.02e-111 | 37 | 739 | 4 | 707 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 1.05e-109 | 37 | 739 | 4 | 707 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 3AC0_A | 3.75e-109 | 36 | 742 | 5 | 833 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXS8 | 3.04e-150 | 15 | 740 | 24 | 740 | Beta-glucosidase BoGH3A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02644 PE=1 SV=1 |
| P14002 | 3.03e-138 | 36 | 745 | 4 | 668 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| P27034 | 2.03e-121 | 37 | 743 | 2 | 808 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| A1CA51 | 9.76e-120 | 36 | 737 | 5 | 822 | Probable beta-glucosidase I OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=bglI PE=3 SV=1 |
| B0Y3M6 | 1.37e-119 | 36 | 737 | 5 | 822 | Probable beta-glucosidase I OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bglI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
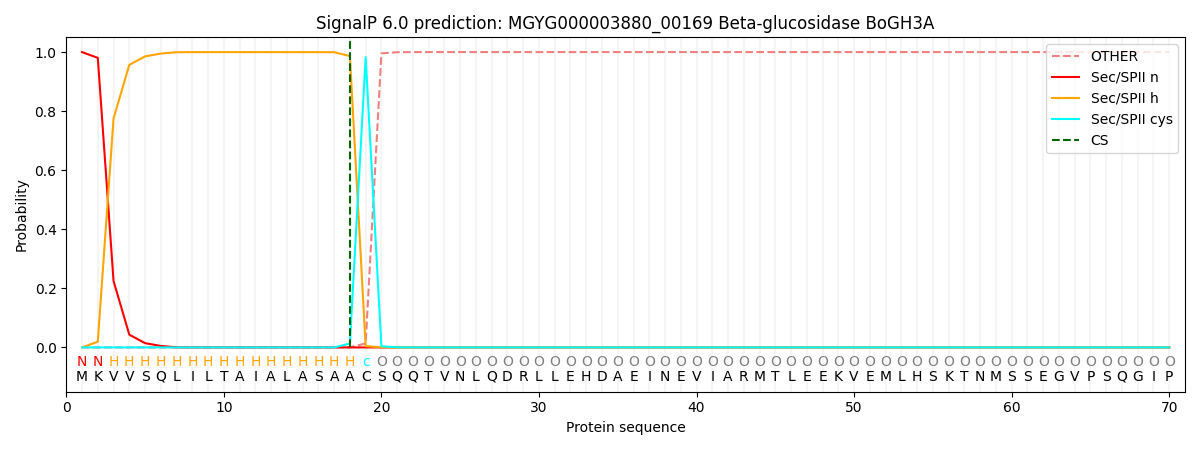
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000050 | 1.000013 | 0.000000 | 0.000000 | 0.000000 |
