You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003904_00099
You are here: Home > Sequence: MGYG000003904_00099
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
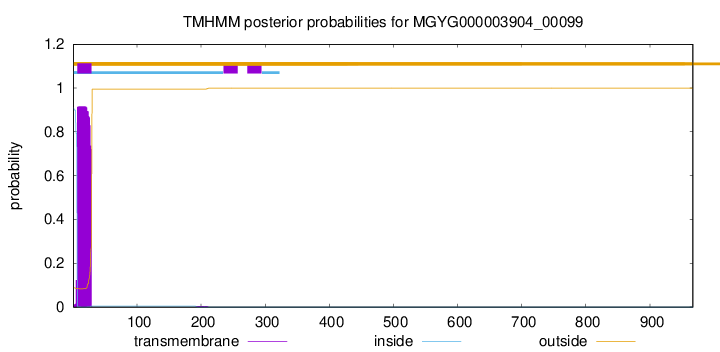
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000003904_00099 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2374; End: 5277 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 318 | 608 | 3.3e-28 | 0.7238979118329466 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd09621 | CBM9_like_5 | 2.17e-55 | 778 | 966 | 1 | 188 | DOMON-like type 9 carbohydrate binding module. Family 9 carbohydrate-binding modules (CBM9) play a role in the microbial degradation of cellulose and hemicellulose (materials found in plants). The domain has previously been called cellulose-binding domain. The polysaccharide binding sites of CBMs with available 3D structure have been found to be either flat surfaces with interactions formed by predominantly aromatic residues (tryptophan and tyrosine), or extended shallow grooves. CBM9 domains found in this uncharacterized heterogeneous subfamily are often located at the C-terminus of longer proteins and may co-occur with various other functional domains such as glycosyl hydrolases. The CBM9 module in these architectures may be involved in binding to carbohydrates. |
| COG3693 | XynA | 9.54e-08 | 309 | 446 | 69 | 204 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 1.30e-07 | 319 | 446 | 74 | 206 | Glycosyl hydrolases family 39. |
| smart00633 | Glyco_10 | 2.06e-06 | 309 | 431 | 3 | 126 | Glycosyl hydrolase family 10. |
| pfam02449 | Glyco_hydro_42 | 1.02e-05 | 277 | 417 | 1 | 153 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44759.1 | 0.0 | 2 | 967 | 1 | 965 |
| AVM47149.1 | 8.59e-194 | 25 | 967 | 350 | 1307 |
| AVM46276.1 | 3.26e-186 | 22 | 967 | 183 | 1137 |
| AHF94342.1 | 1.48e-156 | 145 | 967 | 201 | 1033 |
| AVM45104.1 | 6.43e-122 | 64 | 965 | 217 | 1133 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 1.85e-23 | 219 | 651 | 57 | 493 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
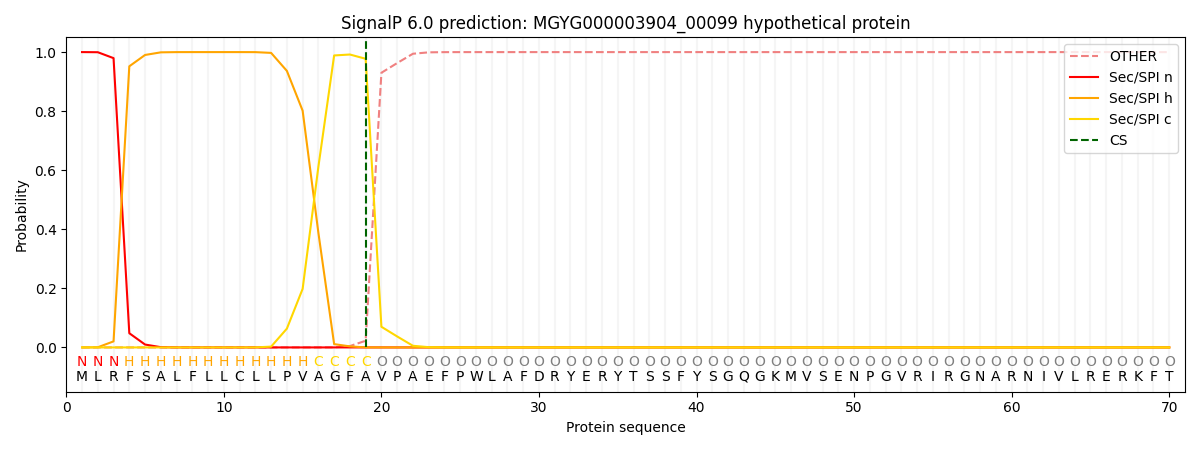
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000227 | 0.999158 | 0.000157 | 0.000161 | 0.000149 | 0.000138 |