You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003904_00932
You are here: Home > Sequence: MGYG000003904_00932
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000003904_00932 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1502; End: 3367 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 69 | 212 | 2.3e-23 | 0.37587006960556846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 7.90e-09 | 55 | 180 | 65 | 200 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| smart00633 | Glyco_10 | 2.28e-08 | 59 | 171 | 3 | 126 | Glycosyl hydrolase family 10. |
| pfam01301 | Glyco_hydro_35 | 0.003 | 29 | 238 | 17 | 230 | Glycosyl hydrolases family 35. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 2.51e-140 | 11 | 618 | 12 | 619 |
| QDU57372.1 | 6.22e-89 | 22 | 585 | 28 | 595 |
| QXQ15225.1 | 1.97e-30 | 32 | 245 | 58 | 291 |
| ACZ43011.1 | 1.09e-29 | 22 | 240 | 302 | 527 |
| QUL56627.1 | 4.78e-29 | 21 | 240 | 384 | 608 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 3.58e-11 | 25 | 243 | 107 | 345 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4ZN2_A | 4.19e-11 | 25 | 243 | 22 | 255 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 4.19e-11 | 25 | 243 | 22 | 255 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 1NQ6_A | 2.92e-08 | 56 | 178 | 45 | 176 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 5M0K_A | 1.58e-06 | 63 | 259 | 89 | 290 | CRYSTALSTRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena],5M0K_B CRYSTAL STRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
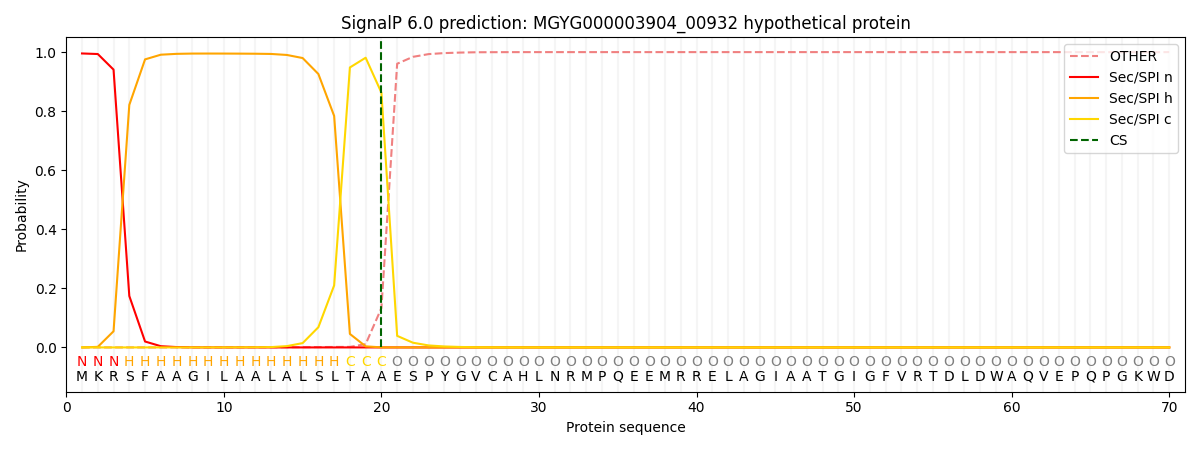
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000715 | 0.993174 | 0.005530 | 0.000218 | 0.000183 | 0.000163 |
