You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003912_00940
You are here: Home > Sequence: MGYG000003912_00940
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000435635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000435635 | |||||||||||
| CAZyme ID | MGYG000003912_00940 | |||||||||||
| CAZy Family | CE19 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 123867; End: 125015 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE19 | 55 | 380 | 8.2e-27 | 0.8433734939759037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0412 | DLH | 2.46e-14 | 108 | 341 | 13 | 182 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam12715 | Abhydrolase_7 | 6.23e-10 | 55 | 340 | 46 | 326 | Abhydrolase family. This is a family of probable bacterial abhydrolases. |
| COG1506 | DAP2 | 5.36e-07 | 165 | 332 | 400 | 567 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam01738 | DLH | 1.56e-05 | 110 | 267 | 1 | 121 | Dienelactone hydrolase family. |
| pfam00326 | Peptidase_S9 | 7.93e-05 | 174 | 271 | 9 | 94 | Prolyl oligopeptidase family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37157.1 | 1.81e-121 | 20 | 377 | 368 | 723 |
| QEG26086.1 | 6.47e-26 | 37 | 382 | 9 | 329 |
| VTS00541.1 | 6.47e-26 | 37 | 382 | 9 | 329 |
| AWM40652.1 | 6.47e-26 | 37 | 382 | 9 | 329 |
| BCS33230.1 | 2.24e-22 | 49 | 378 | 43 | 333 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34973 | 1.48e-29 | 60 | 382 | 2 | 299 | Putative hydrolase YtaP OS=Bacillus subtilis (strain 168) OX=224308 GN=ytaP PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
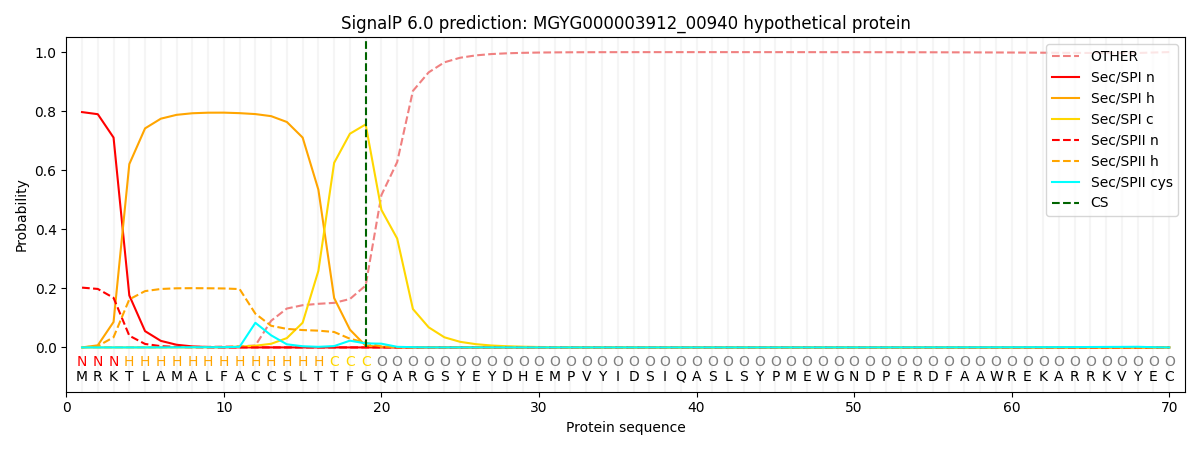
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002014 | 0.784065 | 0.212791 | 0.000461 | 0.000337 | 0.000296 |
