You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003914_01661
You are here: Home > Sequence: MGYG000003914_01661
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Sutterella; | |||||||||||
| CAZyme ID | MGYG000003914_01661 | |||||||||||
| CAZy Family | GH102 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2808; End: 3986 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH102 | 130 | 285 | 2.3e-59 | 0.9936305732484076 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2821 | MltA | 2.11e-106 | 45 | 388 | 33 | 371 | Membrane-bound lytic murein transglycosylase [Cell wall/membrane/envelope biogenesis]. |
| pfam03562 | MltA | 1.59e-104 | 53 | 284 | 4 | 230 | MltA specific insert domain. This beta barrel domain is found inserted in the MltA a murein degrading transglycosylase enzyme. This domain may be involved in peptidoglycan binding. |
| cd14668 | mlta_B | 7.28e-82 | 130 | 287 | 1 | 159 | Domain B insert of mltA_like lytic transglycosylases. Escherichia coli MltA is a membrane-bound lytic transglycosylase comprised of two domains separated by a large groove, where the peptidoglycan strand binds. Domain A is made up of an N-terminal and a C-terminal portion, which correspond to the 3D domain, named for 3 conserved aspartate residues. Domain B is inserted within the linear sequence of domain A. MltA is distinct from other bacterial lytic transglycosylases (LTs), which are similar to each other. Escherichia coli peptidoglycan lytic transglycosylase (LT) initiates cell wall recycling in response to damage, during bacterial fission, and cleaves peptidoglycan (PG) to create functional spaces in its wall. PG chains (also known as murein), the major components of the bacterial cell wall, are comprised of alternating beta-1-4-linked N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), and lytic transglycosylases cleave this beta-1-4 bond. Typically, peptidoglycan lytic transglycosylases (LT) are exolytic, releasing Metabolite 1 (GlcNAc-anhMurNAc-L-Ala-D-Glu-m-Dap-D-Ala-D-Ala) from the ends of the PG strands. In contrast, MltE is endolytic , cleaving in the middle of PG strands, with further processing to Metabolite 1 accomplished by other LTs. In E. coli, there are six membrane-bound LTs: MltA-MltF and soluble Slt70. Slt35 is a soluble fragment cleaved from MltB. Bacterial LTs are classified in 4 families: Family 1 includes slt70 MltC-MltF, Family 2 includes MltA, Family 3 includes MltB, and Family 4 of bacteriophage origin. While most of the LT family members are similar in structure and sequence with a lysozyme-like fold, Family 2 (including mltA) is distinct. |
| smart00925 | MltA | 6.64e-75 | 129 | 284 | 1 | 152 | MltA specific insert domain. This beta barrel domain is found inserted in the MltA a murein degrading transglycosylase enzyme. This domain may be involved in peptidoglycan binding. |
| PRK11162 | mltA | 8.20e-59 | 121 | 380 | 117 | 354 | murein transglycosylase A; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBF22454.1 | 5.90e-198 | 7 | 391 | 6 | 394 |
| QDA55021.1 | 1.24e-175 | 9 | 389 | 20 | 393 |
| QQS89395.1 | 1.14e-164 | 39 | 388 | 47 | 394 |
| ANU66434.1 | 4.12e-149 | 10 | 391 | 12 | 400 |
| QQQ97583.1 | 4.12e-149 | 10 | 391 | 12 | 400 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7ESJ_A | 5.76e-84 | 23 | 385 | 9 | 362 | ChainA, membrane-bound lytic murein transglycosylase A [Acinetobacter baumannii],7ESJ_B Chain B, membrane-bound lytic murein transglycosylase A [Acinetobacter baumannii] |
| 2G5D_A | 8.38e-83 | 39 | 388 | 42 | 419 | Crystalstructure of MltA from Neisseria gonorrhoeae Monoclinic form [Neisseria gonorrhoeae FA 1090] |
| 2G6G_A | 8.16e-80 | 39 | 388 | 42 | 419 | Crystalstructure of MltA from Neisseria gonorrhoeae [Neisseria gonorrhoeae FA 1090] |
| 6QK4_B | 3.33e-78 | 61 | 383 | 47 | 347 | Lytictransglycosylase, LtgG, of Burkholderia pseudomallei. [Burkholderia pseudomallei] |
| 3CZB_A | 1.03e-51 | 41 | 383 | 9 | 342 | Crystalstructure of putative transglycosylase from Caulobacter crescentus [Caulobacter vibrioides CB15],3CZB_B Crystal structure of putative transglycosylase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0A935 | 3.63e-36 | 123 | 380 | 119 | 353 | Membrane-bound lytic murein transglycosylase A OS=Escherichia coli (strain K12) OX=83333 GN=mltA PE=1 SV=1 |
| P0A936 | 3.63e-36 | 123 | 380 | 119 | 353 | Membrane-bound lytic murein transglycosylase A OS=Escherichia coli O157:H7 OX=83334 GN=mltA PE=3 SV=1 |
| Q8K9A7 | 7.42e-27 | 122 | 380 | 126 | 364 | Membrane-bound lytic murein transglycosylase A homolog OS=Buchnera aphidicola subsp. Schizaphis graminum (strain Sg) OX=198804 GN=mltA PE=3 SV=1 |
| Q9KPQ4 | 5.16e-26 | 103 | 380 | 97 | 349 | Membrane-bound lytic murein transglycosylase A OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=mltA PE=3 SV=1 |
| P57531 | 1.84e-23 | 122 | 380 | 118 | 356 | Membrane-bound lytic murein transglycosylase A homolog OS=Buchnera aphidicola subsp. Acyrthosiphon pisum (strain APS) OX=107806 GN=mltA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
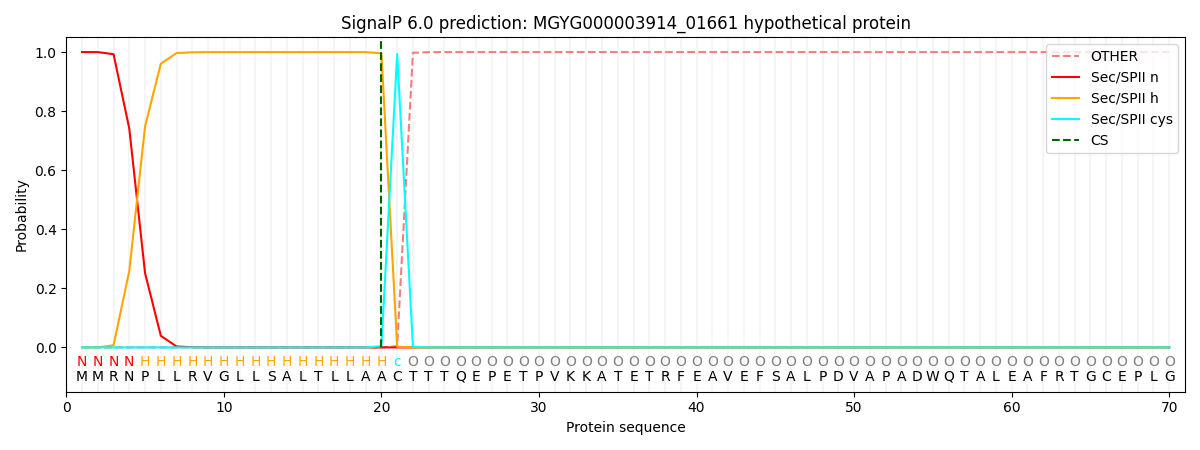
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000047 | 0.000000 | 0.000000 | 0.000000 |
