You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003948_00442
You are here: Home > Sequence: MGYG000003948_00442
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; | |||||||||||
| CAZyme ID | MGYG000003948_00442 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15833; End: 17182 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 62 | 249 | 3.5e-17 | 0.45614035087719296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 2.18e-33 | 61 | 418 | 1 | 342 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 4.71e-09 | 64 | 193 | 1 | 118 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDN31231.1 | 9.94e-226 | 4 | 448 | 3 | 451 |
| BCG53919.1 | 9.93e-223 | 3 | 449 | 4 | 457 |
| QUT52998.1 | 3.89e-222 | 3 | 449 | 4 | 455 |
| BBK93797.1 | 1.11e-221 | 3 | 449 | 4 | 455 |
| QIX65768.1 | 1.11e-221 | 3 | 449 | 4 | 455 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CEA_A | 9.50e-11 | 62 | 286 | 7 | 207 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 3MOI_A | 4.44e-07 | 137 | 278 | 65 | 191 | Thecrystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 [Bordetella bronchiseptica] |
| 3EC7_A | 9.56e-07 | 61 | 276 | 21 | 205 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 6Z3B_B | 9.91e-07 | 109 | 307 | 34 | 220 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
| 6Z3B_A | 1.00e-06 | 109 | 307 | 36 | 222 | Lowresolution structure of RgNanOx [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01S58 | 5.24e-08 | 4 | 250 | 2 | 229 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| A8H2K3 | 5.18e-07 | 5 | 233 | 2 | 217 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
| A6WQ58 | 2.13e-06 | 20 | 233 | 21 | 219 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS185) OX=402882 GN=Shew185_2813 PE=3 SV=1 |
| A3D6B7 | 4.94e-06 | 42 | 233 | 36 | 219 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS155 / ATCC BAA-1091) OX=325240 GN=Sbal_2793 PE=3 SV=1 |
| A9KUT1 | 4.94e-06 | 42 | 233 | 36 | 219 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS195) OX=399599 GN=Sbal195_2933 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
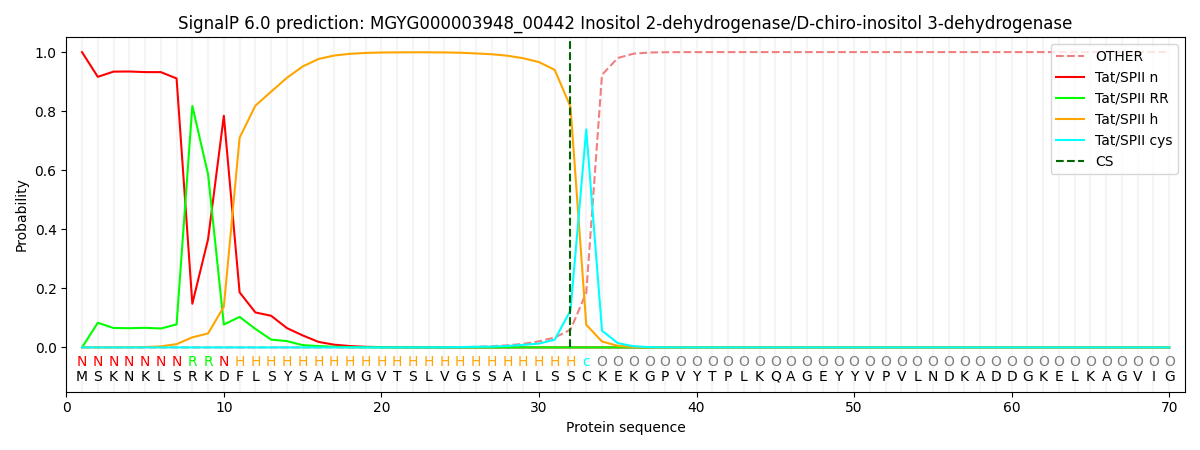
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000004 | 0.000206 | 0.999806 | 0.000000 |
