You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003952_01190
You are here: Home > Sequence: MGYG000003952_01190
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900021155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900021155 | |||||||||||
| CAZyme ID | MGYG000003952_01190 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58688; End: 61315 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 528 | 797 | 4.2e-57 | 0.8316831683168316 |
| GH28 | 141 | 476 | 1.9e-48 | 0.9015384615384615 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.71e-61 | 30 | 478 | 4 | 482 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 1.52e-36 | 528 | 758 | 2 | 230 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.21e-17 | 624 | 858 | 120 | 346 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| PLN02188 | PLN02188 | 5.74e-13 | 111 | 489 | 36 | 391 | polygalacturonase/glycoside hydrolase family protein |
| pfam00295 | Glyco_hydro_28 | 2.75e-12 | 213 | 473 | 52 | 303 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARK08556.1 | 7.10e-95 | 524 | 862 | 23 | 364 |
| AFN73753.1 | 2.92e-94 | 525 | 859 | 36 | 366 |
| AFD07539.1 | 9.58e-94 | 526 | 873 | 50 | 401 |
| AHM61644.1 | 2.11e-93 | 528 | 859 | 30 | 365 |
| QCR22725.1 | 3.72e-93 | 528 | 865 | 28 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q78_A | 3.36e-74 | 513 | 862 | 46 | 406 | Thestructure of GH26C from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q78_B The structure of GH26C from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
| 4ZXO_A | 2.44e-69 | 530 | 859 | 7 | 334 | Thestructure of a GH26 beta-mannanase from Bacteroides ovatus, BoMan26A. [Bacteroides ovatus] |
| 2VX4_A | 3.46e-62 | 503 | 859 | 7 | 386 | CellvibrioJaponicus Mannanase Cjman26c Native Form [Cellvibrio japonicus],2VX6_A CELLVIBRIO JAPONICUS MANNANASE CJMAN26C Gal1Man4-BOUND FORM [Cellvibrio japonicus] |
| 2VX5_A | 3.46e-62 | 503 | 859 | 7 | 386 | CellvibrioJaponicus Mannanase Cjman26c Mannose-Bound Form [Cellvibrio japonicus] |
| 4CD4_A | 6.34e-62 | 503 | 859 | 30 | 409 | Thestructure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG [Cellvibrio japonicus Ueda107],4CD5_A The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm [Cellvibrio japonicus Ueda107] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 1.25e-46 | 525 | 835 | 48 | 366 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 2.30e-42 | 526 | 862 | 42 | 396 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P15922 | 4.67e-32 | 16 | 517 | 26 | 602 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| O05512 | 8.58e-16 | 574 | 759 | 79 | 269 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 3.71e-14 | 616 | 759 | 121 | 267 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
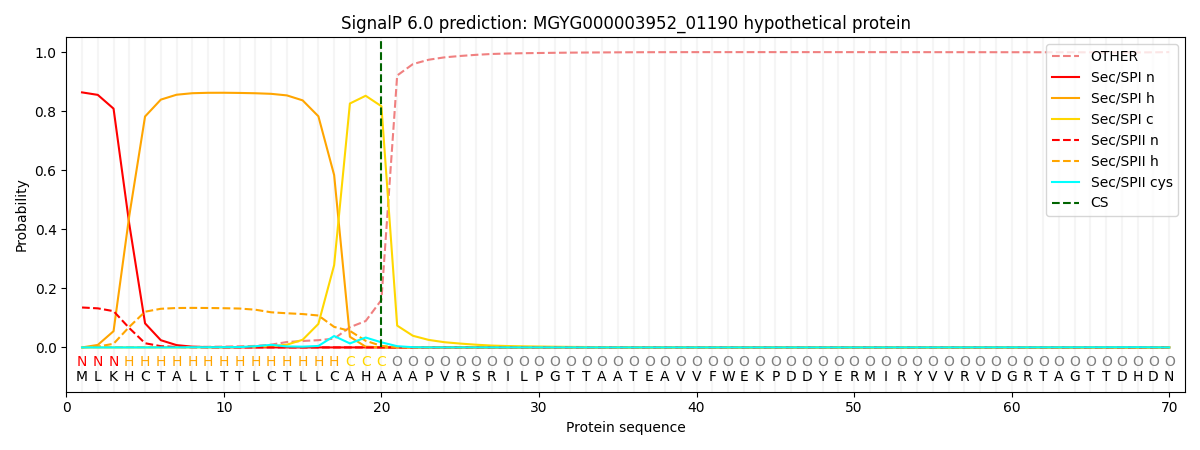
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002267 | 0.856755 | 0.140106 | 0.000361 | 0.000241 | 0.000231 |
