You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003957_00186
You are here: Home > Sequence: MGYG000003957_00186
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp014287585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp014287585 | |||||||||||
| CAZyme ID | MGYG000003957_00186 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 105589; End: 107940 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 27 | 540 | 2.4e-67 | 0.9867172675521821 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00987 | PDZ_serine_protease | 8.45e-14 | 691 | 782 | 2 | 90 | PDZ domain of trypsin-like serine proteases, such as DegP/HtrA, which are oligomeric proteins involved in heat-shock response, chaperone function, and apoptosis. May be responsible for substrate recognition and/or binding, as most PDZ domains bind C-terminal polypeptides, though binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. In this subfamily of protease-associated PDZ domains a C-terminal beta-strand forms the peptide-binding groove base, a circular permutation with respect to PDZ domains found in Eumetazoan signaling proteins. |
| pfam13229 | Beta_helix | 7.68e-09 | 312 | 463 | 1 | 136 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| cd00136 | PDZ | 3.48e-06 | 690 | 777 | 1 | 64 | PDZ domain, also called DHR (Dlg homologous region) or GLGF (after a conserved sequence motif). Many PDZ domains bind C-terminal polypeptides, though binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. Heterodimerization through PDZ-PDZ domain interactions adds to the domain's versatility, and PDZ domain-mediated interactions may be modulated dynamically through target phosphorylation. Some PDZ domains play a role in scaffolding supramolecular complexes. PDZ domains are found in diverse signaling proteins in bacteria, archebacteria, and eurkayotes. This CD contains two distinct structural subgroups with either a N- or C-terminal beta-strand forming the peptide-binding groove base. The circular permutation placing the strand on the N-terminus appears to be found in Eumetazoa only, while the C-terminal variant is found in all three kingdoms of life, and seems to co-occur with protease domains. PDZ domains have been named after PSD95(post synaptic density protein), DlgA (Drosophila disc large tumor suppressor), and ZO1, a mammalian tight junction protein. |
| pfam17820 | PDZ_6 | 2.63e-05 | 717 | 759 | 1 | 43 | PDZ domain. This entry represents the PDZ domain from a wide variety of proteins. |
| pfam13229 | Beta_helix | 5.20e-05 | 335 | 564 | 1 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWW32784.1 | 2.62e-210 | 12 | 765 | 15 | 782 |
| BAU63437.1 | 2.32e-51 | 26 | 660 | 56 | 728 |
| AFZ35556.1 | 1.90e-44 | 28 | 660 | 49 | 710 |
| QIP13872.1 | 2.65e-37 | 14 | 568 | 13 | 545 |
| ALJ04165.1 | 8.27e-36 | 15 | 660 | 12 | 670 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 3.28e-18 | 27 | 449 | 27 | 501 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
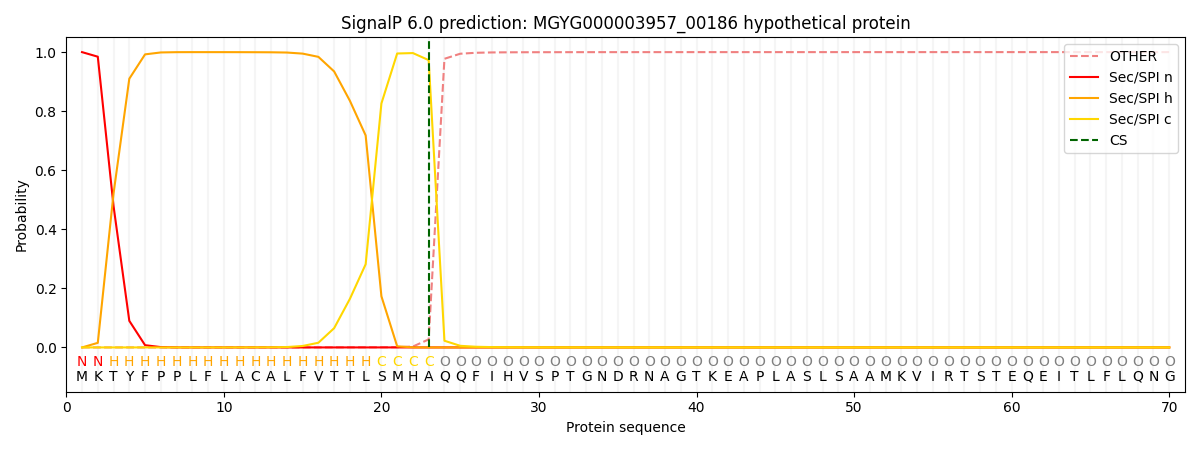
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000575 | 0.998350 | 0.000441 | 0.000206 | 0.000190 | 0.000192 |
