You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003957_01209
Basic Information
help
| Species |
Parabacteroides sp014287585
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp014287585
|
| CAZyme ID |
MGYG000003957_01209
|
| CAZy Family |
PL27 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003957 |
4724401 |
MAG |
United Kingdom |
Europe |
|
| Gene Location |
Start: 56133;
End: 58211
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| PL27 |
62 |
686 |
3.2e-244 |
0.9967213114754099 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam18951
|
DUF5695 |
1.38e-07 |
310 |
532 |
324 |
590 |
Family of unknown function (DUF5695). This is a family of unknown function mainly found in fungi and bacteria. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5NO8_A
|
0.0 |
8 |
690 |
15 |
692 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
| 5NOK_A
|
0.0 |
8 |
690 |
15 |
692 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
This protein is predicted as LIPO
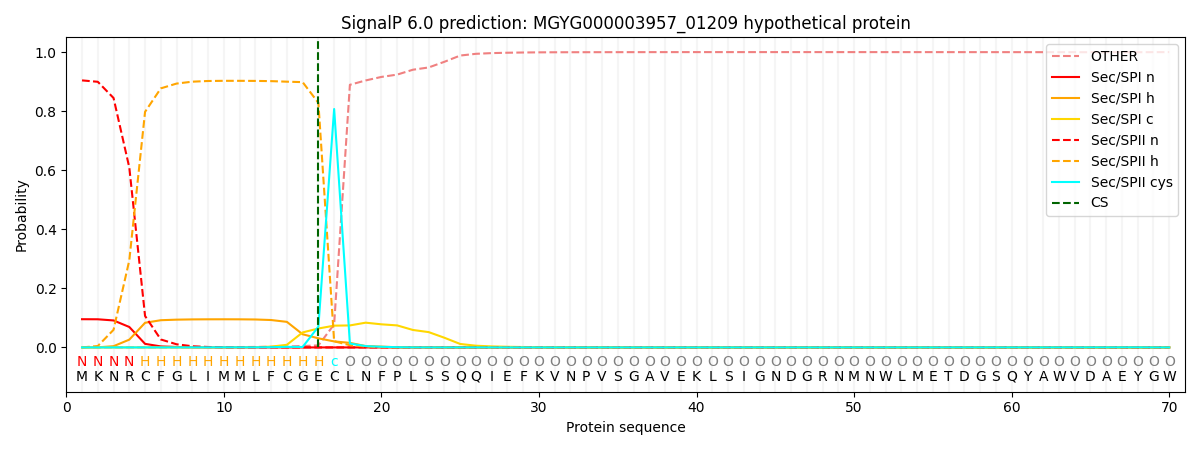
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000479
|
0.093508
|
0.905903
|
0.000042
|
0.000044
|
0.000039
|
There is no transmembrane helices in MGYG000003957_01209.

