You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003958_00289
You are here: Home > Sequence: MGYG000003958_00289
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
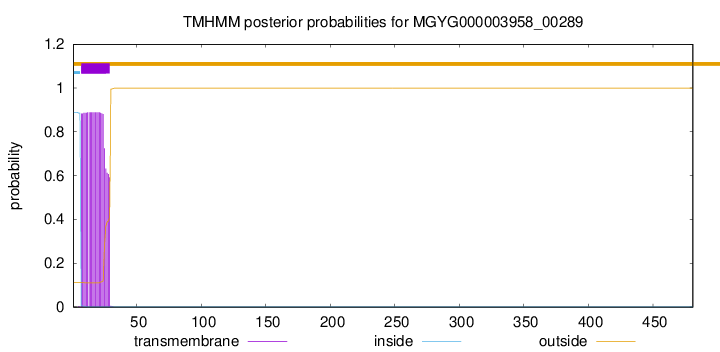
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900545715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900545715 | |||||||||||
| CAZyme ID | MGYG000003958_00289 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 344876; End: 346321 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 67 | 327 | 1e-20 | 0.6836363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11309 | 14-3-3_fungi | 2.57e-04 | 196 | 305 | 59 | 168 | Fungal 14-3-3 protein domain. This family containing fungal 14-3-3 domains includes the yeasts Saccharomyces cerevisiae (BMH1 and BMH2) and Schizosaccharomyces pombe (rad24 and rad25) isoforms. They possess distinctively variant C-terminal segments that differentiate them from the mammalian isoforms; the C-terminus is longer and BMH1/2 isoforms contain polyglutamine (polyQ) sequences of unknown function. The C-terminal segments of yeast 14-3-3 isoforms may thus behave in a different manner compared to the higher eukaryote isoforms. Yeast 14-3-3 proteins bind to numerous proteins involved in a variety of yeast cellular processes making them excellent model organisms for elucidating the function of the 14-3-3 protein family. BMH1 and BMH2 are positive regulators of rapamycin-sensitive signaling via TOR kinases while they play an inhibitory role in Rtg3p-dependent transcription involved in retrograde signaling. 14-3-3 domains are an essential part of 14-3-3 proteins, a ubiquitous class of regulatory, phosphoserine/threonine-binding proteins found in all eukaryotic cells, including yeast, protozoa and mammalian cells. |
| pfam00150 | Cellulase | 0.008 | 79 | 271 | 33 | 197 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL08912.1 | 7.72e-114 | 28 | 480 | 32 | 469 |
| BBL11704.1 | 7.72e-114 | 28 | 480 | 32 | 469 |
| BBL00987.1 | 7.72e-114 | 28 | 480 | 32 | 469 |
| AEI51976.1 | 1.66e-111 | 26 | 479 | 27 | 474 |
| AXE20299.1 | 2.10e-109 | 28 | 479 | 29 | 474 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
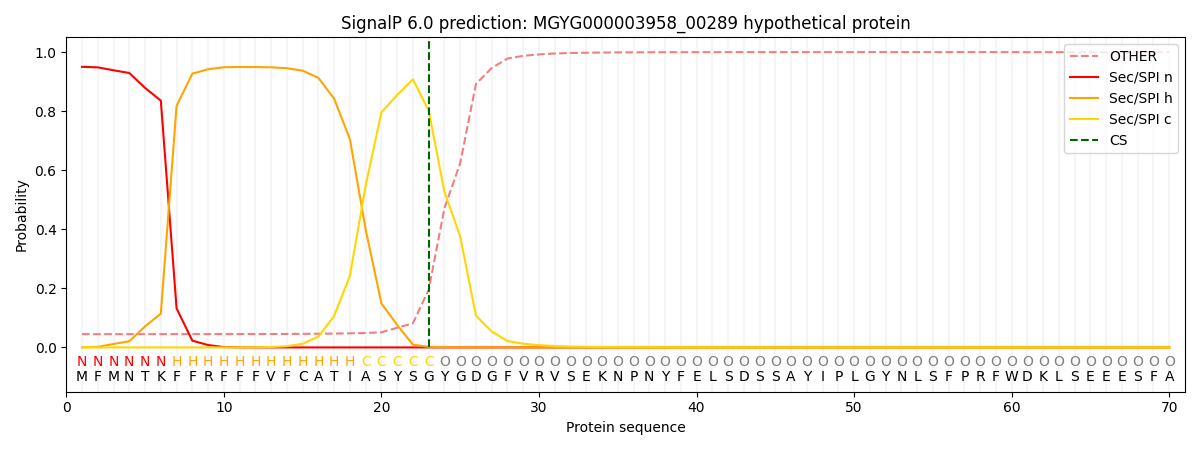
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.054101 | 0.938592 | 0.006300 | 0.000310 | 0.000311 | 0.000342 |