You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003967_01324
You are here: Home > Sequence: MGYG000003967_01324
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
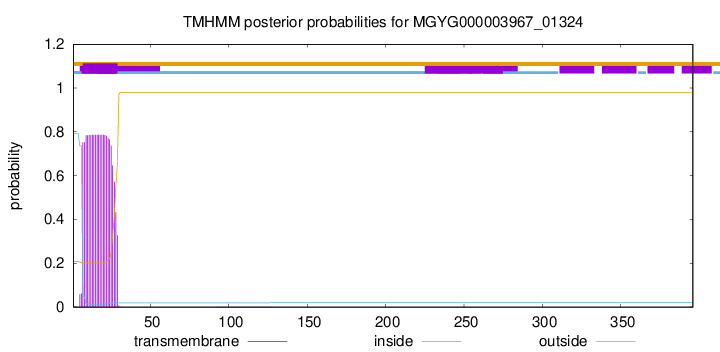
TMHMM annotations
Basic Information help
| Species | UMGS1338 sp900550805 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; QALW01; UMGS1338; UMGS1338 sp900550805 | |||||||||||
| CAZyme ID | MGYG000003967_01324 | |||||||||||
| CAZy Family | CE2 | |||||||||||
| CAZyme Description | Cellulase/esterase CelE | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6948; End: 8138 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 193 | 386 | 3.1e-50 | 0.9904306220095693 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01831 | Endoglucanase_E_like | 4.70e-31 | 193 | 386 | 2 | 168 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| pfam13472 | Lipase_GDSL_2 | 8.57e-12 | 195 | 328 | 1 | 104 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 2.98e-09 | 193 | 384 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd01825 | SGNH_hydrolase_peri1 | 7.36e-06 | 261 | 389 | 32 | 188 | SGNH_peri1; putative periplasmic member of the SGNH-family of hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| cd01827 | sialate_O-acetylesterase_like1 | 9.67e-06 | 196 | 387 | 6 | 188 | sialate O-acetylesterase_like family of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16806.1 | 1.45e-31 | 131 | 393 | 158 | 432 |
| ADC90443.1 | 1.52e-31 | 59 | 392 | 38 | 395 |
| CBL33310.1 | 8.41e-31 | 72 | 391 | 9 | 356 |
| CBK96966.1 | 1.61e-30 | 72 | 391 | 9 | 356 |
| AAA23224.1 | 7.14e-29 | 73 | 391 | 488 | 813 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WAO_A | 8.24e-32 | 73 | 391 | 7 | 332 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 2WAB_A | 2.19e-31 | 73 | 391 | 7 | 332 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 3U37_A | 2.57e-22 | 190 | 391 | 165 | 405 | AnAcetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_B An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_C An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_D An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_E An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_F An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_G An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_H An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 4DEV_A | 4.01e-21 | 190 | 391 | 165 | 405 | AnAcetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_B An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_C An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_D An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_E An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_F An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_G An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_H An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 2W9X_A | 3.90e-15 | 124 | 391 | 78 | 358 | Theactive site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus],2W9X_B The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10477 | 1.43e-29 | 73 | 391 | 488 | 813 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| B3PDE5 | 2.05e-14 | 124 | 391 | 78 | 358 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ce2C PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
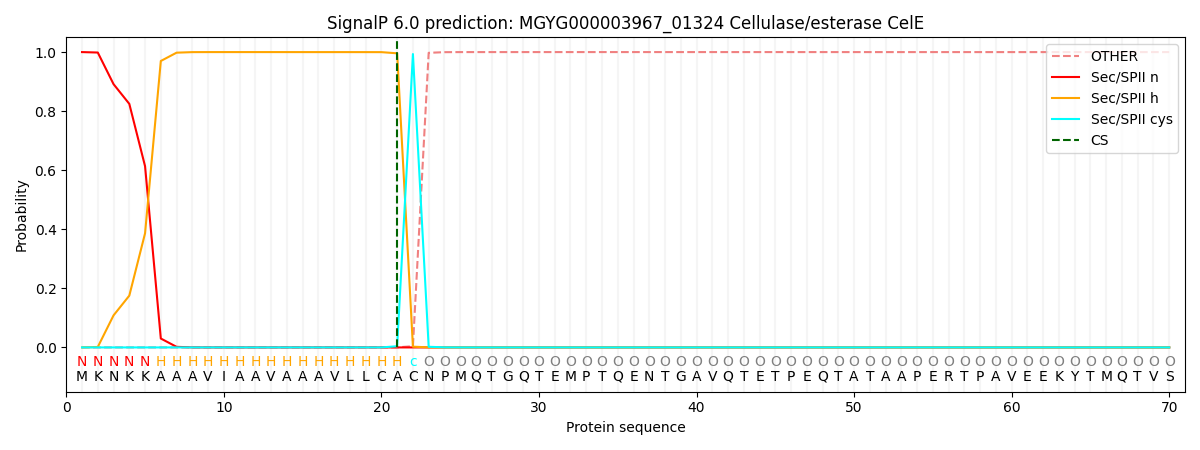
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |