You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003984_01271
You are here: Home > Sequence: MGYG000003984_01271
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
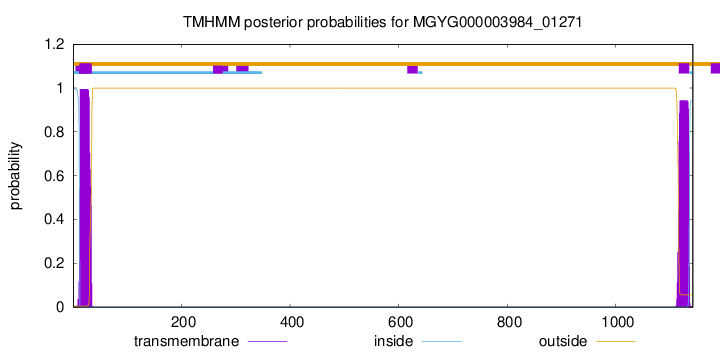
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900752395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900752395 | |||||||||||
| CAZyme ID | MGYG000003984_01271 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30249; End: 33677 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 423 | 967 | 1.1e-149 | 0.9405204460966543 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13320 | DUF4091 | 1.16e-16 | 864 | 935 | 1 | 65 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| COG5492 | YjdB | 3.63e-08 | 319 | 404 | 177 | 261 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 5.63e-07 | 324 | 390 | 1 | 66 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| smart00635 | BID_2 | 3.06e-05 | 322 | 390 | 1 | 71 | Bacterial Ig-like domain 2. |
| TIGR01167 | LPXTG_anchor | 0.004 | 1109 | 1138 | 3 | 32 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUD71731.1 | 4.30e-223 | 39 | 1002 | 49 | 999 |
| AYE33261.1 | 3.23e-200 | 171 | 995 | 34 | 838 |
| QAS61432.1 | 3.23e-200 | 171 | 995 | 34 | 838 |
| QBJ76609.1 | 1.00e-193 | 410 | 999 | 6 | 581 |
| ATD58699.1 | 1.00e-193 | 410 | 999 | 6 | 581 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FQE_A | 2.21e-178 | 411 | 995 | 30 | 600 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQE_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FR0_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5FQG_A | 3.41e-177 | 411 | 995 | 30 | 600 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQH_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5L7V_A | 8.44e-75 | 439 | 935 | 53 | 516 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 1.21e-74 | 439 | 935 | 68 | 531 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR5 | 2.31e-06 | 78 | 311 | 869 | 1070 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
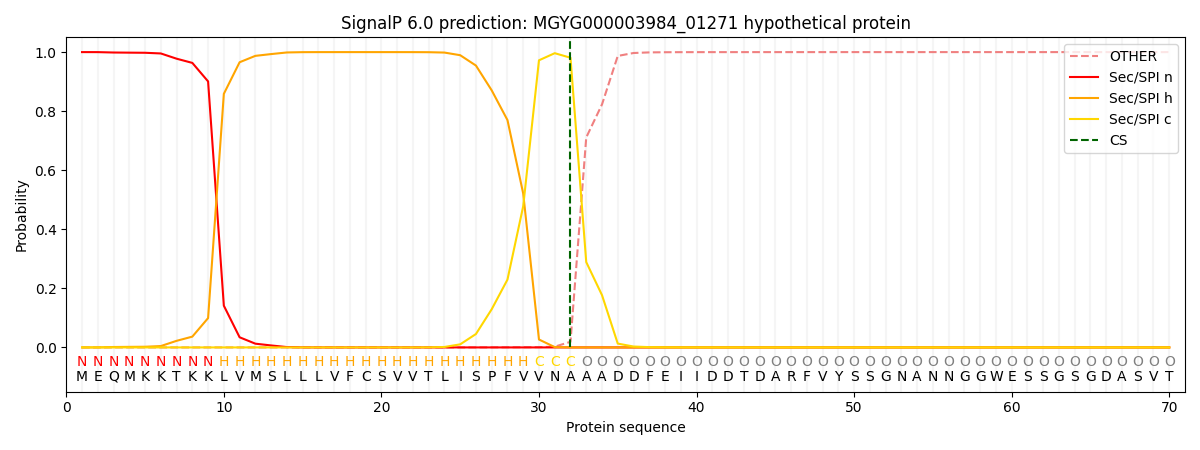
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000341 | 0.998923 | 0.000215 | 0.000183 | 0.000163 | 0.000145 |