You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003997_00231
You are here: Home > Sequence: MGYG000003997_00231
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM12650 sp900761725 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; HGM12650; HGM12650 sp900761725 | |||||||||||
| CAZyme ID | MGYG000003997_00231 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 252575; End: 254497 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 105 | 328 | 2.3e-26 | 0.8766519823788547 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0657 | Aes | 2.65e-12 | 405 | 635 | 48 | 307 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 1.12e-10 | 459 | 617 | 6 | 207 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| COG2382 | Fes | 1.24e-07 | 105 | 251 | 83 | 202 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00326 | Peptidase_S9 | 3.38e-05 | 517 | 618 | 61 | 190 | Prolyl oligopeptidase family. |
| COG0400 | YpfH | 0.001 | 446 | 629 | 13 | 202 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADA67819.1 | 1.88e-112 | 60 | 296 | 35 | 273 |
| AGA59284.1 | 2.21e-44 | 68 | 345 | 316 | 596 |
| QSF48012.1 | 2.88e-43 | 64 | 345 | 347 | 630 |
| QLG42560.1 | 9.76e-43 | 68 | 345 | 324 | 603 |
| ACL77751.1 | 1.86e-42 | 104 | 345 | 443 | 691 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GKL_A | 6.11e-40 | 104 | 344 | 53 | 287 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1GKL_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB4_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB4_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB6_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB6_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FJ4_A | 1.82e-37 | 104 | 344 | 39 | 273 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1GKK_A | 2.57e-37 | 104 | 344 | 53 | 287 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1GKK_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],3ZI7_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],3ZI7_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],4BAG_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4BAG_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4H35_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],4H35_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],5FXM_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_AAA Chain AAA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_BBB Chain BBB, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1JJF_A | 1.14e-14 | 106 | 255 | 48 | 177 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 2.80e-14 | 106 | 255 | 48 | 177 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 1.03e-35 | 104 | 344 | 841 | 1075 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P10478 | 5.97e-13 | 106 | 255 | 67 | 196 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
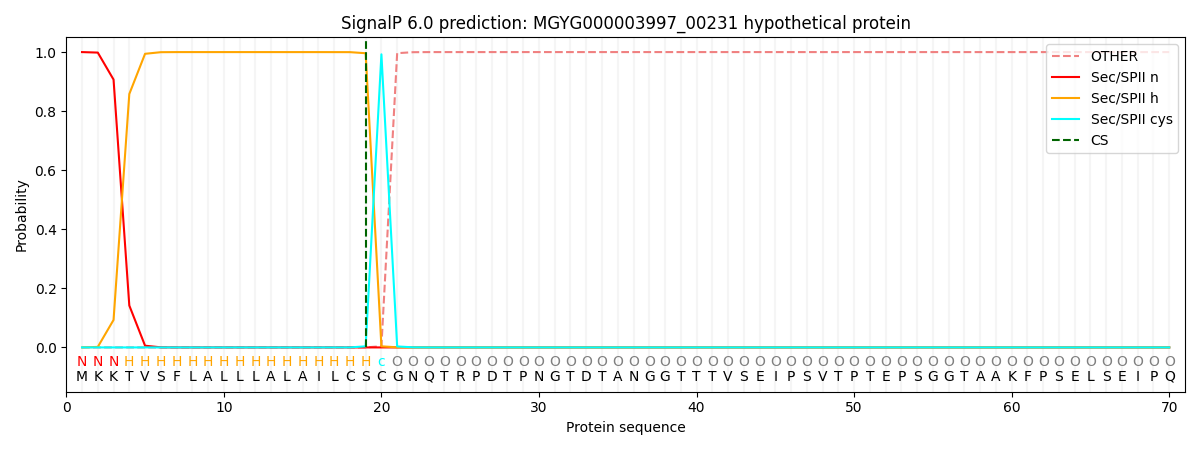
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000041 | 0.000000 | 0.000000 | 0.000000 |
