You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004003_01661
You are here: Home > Sequence: MGYG000004003_01661
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000004003_01661 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36795; End: 38363 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 57 | 509 | 3.3e-136 | 0.9933333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 2.37e-23 | 64 | 473 | 43 | 384 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02055 | Glyco_hydro_30 | 9.84e-08 | 68 | 413 | 1 | 348 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam14587 | Glyco_hydr_30_2 | 4.54e-07 | 56 | 372 | 3 | 353 | O-Glycosyl hydrolase family 30. |
| pfam02057 | Glyco_hydro_59 | 0.010 | 202 | 373 | 110 | 252 | Glycosyl hydrolase family 59. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA52347.1 | 3.43e-287 | 1 | 515 | 1 | 515 |
| QIU93344.1 | 1.75e-211 | 1 | 515 | 1 | 516 |
| ALJ39695.1 | 4.54e-209 | 1 | 515 | 1 | 515 |
| QMW85450.1 | 4.54e-209 | 1 | 515 | 1 | 515 |
| AAO78132.1 | 4.54e-209 | 1 | 515 | 1 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CKQ_A | 6.28e-21 | 179 | 509 | 102 | 407 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4UQA_A | 1.52e-20 | 179 | 509 | 102 | 407 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 3.69e-20 | 179 | 509 | 102 | 407 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
| 4FMV_A | 2.80e-17 | 150 | 512 | 52 | 386 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| 6IUJ_A | 1.43e-09 | 56 | 420 | 23 | 384 | ChainA, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O16580 | 2.11e-09 | 45 | 478 | 75 | 507 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
| P17439 | 4.52e-08 | 44 | 467 | 73 | 494 | Lysosomal acid glucosylceramidase OS=Mus musculus OX=10090 GN=Gba PE=1 SV=1 |
| G5ECR8 | 1.40e-07 | 55 | 420 | 91 | 457 | Putative glucosylceramidase 3 OS=Caenorhabditis elegans OX=6239 GN=gba-3 PE=3 SV=1 |
| Q9UB00 | 5.61e-07 | 50 | 460 | 81 | 490 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
| Q9BDT0 | 5.73e-07 | 44 | 420 | 93 | 473 | Lysosomal acid glucosylceramidase OS=Pan troglodytes OX=9598 GN=GBA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
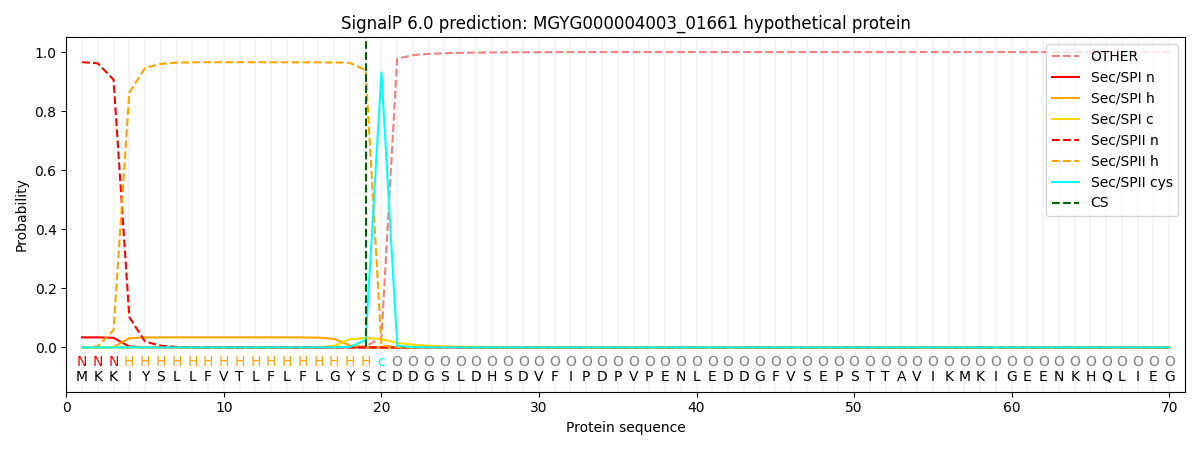
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000170 | 0.033189 | 0.966625 | 0.000014 | 0.000018 | 0.000016 |
